Structural basis for protein glutamylation by the Legionella pseudokinase SidJ
Nature Communications 26 October 2020
10.1038/s41467-021-26429-y
Understanding how two bacterial proteins interact could lead to the development of new drugs to treat pneumonia
By Carolina Araujo Sousa
As a biochemist, Sagar Bhogaraju felt it was a “dream come true” when he started studying Legionella pneumophila during his postdoc. “There are so many interesting proteins there, with such novel functions that you don’t see elsewhere in nature,” he explained. The uniqueness of the species made it the perfect organism to investigate when he started his own group at EMBL Grenoble.
L. pneumophila is a versatile species of bacteria that can infect several different hosts. In humans, it may lead to a mild infection, called Pontiac Fever, or to Legionnaires’ disease, a severe pneumonia. Between 2011 and 2015, 9.3% of Legionnaires’ disease cases in Europe were fatal. Unfortunately, doctors still do not have a reliably effective treatment.
“The treatment regime includes extensive exposure to conventional antibiotics for long periods, which has been shown to cause microbiota changes in the lungs and stomach,” said Dr Bhogaraju, “Complementary treatments are needed.”
Understanding how L. pneumophila infects cells can aid in the development of treatment strategies against it. The Bhogaraju group has recently spent some time studying one particular set of molecular players that are important for the infection process.
When cells need to degrade a protein, they attach a little tag to it, called ubiquitin. A special set of enzymes carry out this tagging process, which is called ubiquitination. In species as different as yeast and humans, ubiquitination occurs in the same three-step manner with a different enzyme carrying out each step. But L. pneumophila has a ‘super’ version of these enzymes – SdeA – which can ubiquitinate other proteins without using the conventional three-step cascade; it’s the only known enzyme in nature to be able to do so on its own.
Ubiquitination of proteins normally occurs in cells and is essential for their survival. When L. pneumophila infects a host, it adds the burden of SdeA-driven toxic ubiquitination on top of the conventional ubiquitination already taking place in the host cell. To survive and multiply within the host, L. pneumophila needs to attack the cell without killing it. Therefore, it needs a way to bring the toxic ubiquitination level down to a tolerable limit.
For this, L. pneumophila has a self-regulatory system – another protein, called SidJ, which reduces SdeA’s activity. It does so by attaching another tag – glutamate – to the SdeA protein itself, a process called glutamylation. But it remains unclear how SidJ and SdeA interact with each other to allow glutamylation to happen. “If we want to, one day, come up with a drug to treat Legionella pneumophila infections, we need to understand the reactions going on between its proteins,” said Michael Adams, a predoc at Bhogaraju group and the first author of a recent paper in Nature communications.
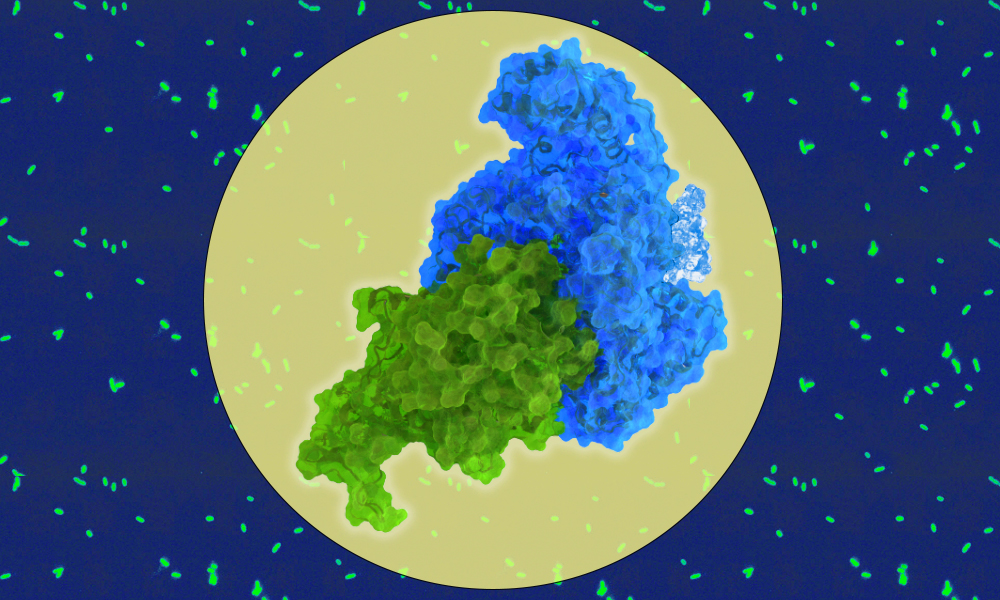
To act in concert, SidJ and SdeA have to fit into each other like two pieces in a jigsaw puzzle. However, given the proteins’ structures, there are two possible ways for this to happen. SidJ has two ‘pockets’ into which SdeA can fit and until now, it was not clear which of these was important for the glutamylation process. Scientists in the Bhogaraju group decided to investigate this, in collaboration with scientists from the Max Planck Institute for Biology of Ageing.
“Using a powerful microscopy method at EMBL Heidelberg, known as cryo-EM, we’ve been able to better understand the structures of both of these molecules and how they interact,” Adams explained.
For this, first, the scientists prepared a solution containing the two proteins and the glutamate tag. Next, the samples were frozen in such a way that the researchers could capture the structure from different angles, ultimately creating a 3D picture of what was happening.
With this technique, the group found that both pockets are important for the interaction of SidJ with SdeA, but the glutamate tag is added to SdeA at only one of them. “It appears that both pockets are necessary and they can both be targeted for drug development,” said Dr Bhogaraju.
With the 3D structure of the SidJ-SdeA-glutamate complex solved, scientists are closer to understanding how these components interact within an infected cell. “One next step is to try to target these proteins, now that we have a lot of information about them,” said Dr Bhogaraju.
Nature Communications 26 October 2020
10.1038/s41467-021-26429-y
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive