
Sagar Bhogaraju
Group Leader
ORCID: 0000-0001-9998-5927
EditStructural biology of ubiquitin signalling

Group Leader
ORCID: 0000-0001-9998-5927
EditUbiquitin is a 76-amino acid polypeptide that is one of the most abundant proteins in eukaryotic cells. It is covalently attached to other proteins in a process known as ubiquitination, which often leads to protein degradation but can also alter the function of the modified protein. Ubiquitination regulates many cellular processes, including protein turnover, DNA repair, cell division, vesicular transport, autophagy and innate immunity. Misregulation of ubiquitination is involved in a number of major human diseases. The attachment of ubiquitin to proteins is carried out by a cascade of three enzymatic reactions involving E1, E2, and E3 enzymes, resulting in the attachment of the ubiquitin C-terminal carboxyl group to a lysine residue in the substrate protein via an isopeptide bond. With over 600 genes coding for E3 ubiquitin ligases in humans, these enzymes make up a significant portion of the human genome. Our group studies the structure, function, and regulation of unique, understudied ubiquitin ligase complexes of human and bacterial origin.
Melanoma-associated antigens (MAGEs)- A family of proteins regulating ubiquitin ligases
The MAGE family of genes encode for around 40 different proteins that share a common MAGE homology domain. Linked to several neurodevelopmental disorders and tumorigenesis, the MAGE proteins form complexes called MAGE-RING ligases (MRLs) with various RING ubiquitin ligases, which target specific cellular substrates for ubiquitination (Lee and Potts, 2017). The precise role of MAGE proteins in the MRL complexes needs to be better understood and it is not clear how these proteins with 40-80% sequence similarity are able to recognize a diverse range of RING proteins. The mechanism by which misregulation or mutations in a MAGE gene leads to human disease remains a mystery in most cases. Our approach to these questions involves carrying out structural, biochemical and functional examinations on several MAGE proteins and associated complexes.
Our latest research (Griffith-Jones et al., 2024) showed how a cancer specific MAGE-A4 interacts with its partner ubiquitin ligase RAD18. This has provided insights into number of characteristics of MAGEs. Two of the major findings are listed below.
1) We found that MAGE-A4 uses a specialized peptide-binding groove for interacting with its partner ligase. This groove seems to be also conserved in other MAGE proteins that we have analyzed indicating that there may be a common mechanism of ligase recognition adopted by MAGE proteins.
2) MAGE-A4, through its binding to RAD18, inhibits the degradative auto-ubiquitination of RAD18 to stabilize it in cells.
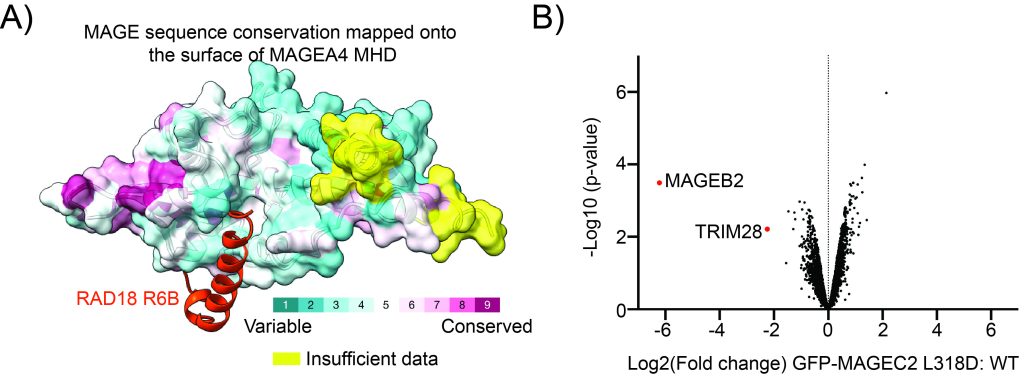
Figure: The ligase-binding cleft in MAGE proteins. A) AlphaFold model of MAGE-A4 bound to a short peptide from the ubiquitin ligase RAD18. MAGE-A4 is depicted in surface representation with sequence conservation amongst MAGE proteins mapped onto the surface. B) Quantitative mass spectrometry data represented in Volcano plot showing that a single point mutation of the equivalent groove in MAGE-C2 results in loss of binding to its partner ubiquitin ligase TRIM28 in HEK cells.
Our ongoing work will build on these findings and in this context, we are also keenly exploring the molecular and cellular mechanisms of other disease relevant MAGE proteins.
An accessible method to identify substrates of ubiquitin ligases
Given the large gaps in the human ubiquitin ligase-substrate pair landscape, there is a need to develop a robust but resource lite methodology to identify the substrates of ubiquitin ligases. we developed a ubiquitin-specific proximity-based labelling method to selectively biotinylate substrates of a given ubiquitin ligase. Our method exploits the proximity of the E3-ligase catalytic domain with respect to ubiquitin observed in the enzymatic intermediate-state structures of E3-E2~Ub. By fusing the biotin ligase BirA and an Avi-tag variant to the candidate E3 ligase and ubiquitin, respectively, we were able to specifically enrich bona fide substrates and potential new substrates of a ligase using a one-step streptavidin pulldown under denaturing conditions. In a recent study, we applied our method (Ub-POD) to three different ubiquitin ligases successfully (Mukhopadhyay et al., 2023) and are currently planning to expand this systematically to more ligases. We also intend to improve the specificity and efficiency of the method further.
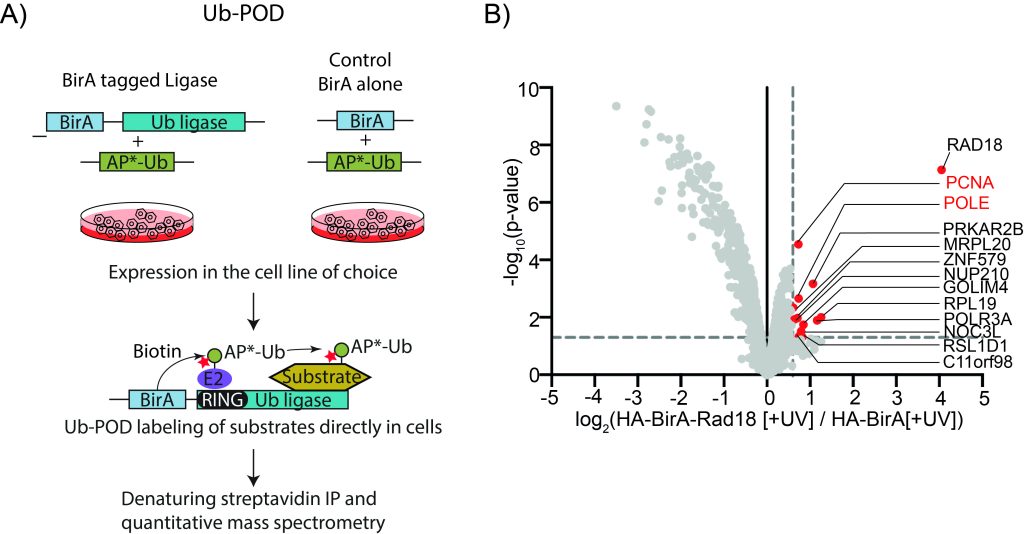
Figure: A) Overview of Ub-POD methodology to identify substrates of ubiquitin ligases. B) Ub-POD applied to the ubiquitin ligase RAD18. Quantitative mass spectrometry data represented in Volcano plot showing the identification of known and potentially novel substrates.
Exploitation of Host Ubiquitination by Legionella pneumophila
Bacteria do not have a ubiquitin system, but some pathogenic bacteria have developed virulence factors (effectors) that can manipulate the host’s ubiquitin system. During an infection, these effectors are secreted into the host cell, allowing the pathogen to evade the host’s immune system and establish a successful environment for replication in the host’s cytoplasm. Legionella bacteria are commonly found in soil and natural water sources and can cause Legionnaires’ disease, a severe form of pneumonia, if inhaled in the form of aerosols. Elderly individuals and those with compromised immune systems are at high risk of fatal complications. During an infection, Legionella secretes over 300 effectors into the host’s cytosol to control various cell-signaling events, including the ubiquitin signaling system. Our research focuses on how these Legionella effectors interact with the host’s ubiquitin system, using tools from structural biology, cell biology, and proteomics.
Our research, along with that of others, uncovered that SdeA, a toxic effector of Legionella, employs a completely different mechanism compared to the standard three enzyme cascade for attaching ubiquitin to its substrates. (Qiu et al., 2016; Bhogaraju et al., 2016). SdeA-mediated reaction results in the attachment of Arginine-42 of ubiquitin to Serines of target substrates through a phosphoribose linker. This unique process of linking ubiquitin to a substrate protein involves the formation of a phosphodiester bond instead of the typical isopeptide bond. It’s still unclear if this distinct form of ubiquitination is carried out by enzymes in other living organisms. We investigated the regulation of this unusual form of ubiquitination by SidJ, a meta-effector of Legionella. SidJ regulates the excessive toxicity caused by SdeA and its paralogues during Legionella infection in a specific and timely manner. Our research found that SidJ partners with host Calmodulin to inhibit the activity of SdeA through ATP-dependent glutamylation. The cryo-electron microscopy (Cryo-EM) structure of SidJ in complex with human Calmodulin revealed how Calmodulin activates SidJ allosterically. This structure also demonstrated that SidJ has a pseudokinase fold with two nucleotide-binding pockets (Bhogaraju et al., 2019). Recently, we used cryo-EM to determine the structure of two catalytic intermediate states of the SidJ/Calmodulin complexed with SdeA (Figure 1). This uncovered a novel mechanism of glutamylation that involves a fleeting self-modification of SidJ through Lysine AMPylation (Adams et al., 2021). In our work on other ubiquitin ligases of legionella, we determined the cryo-EM structure of the ligase LubX in complex with its substrate and another effector of Legionella SidH. Our structure revealed that SidH possesses a tRNA binding site that is important for its celluylar toxicity. We are continuing to study ubiquitin signalling in Legionella, especially in the context of infections, through collaborations.
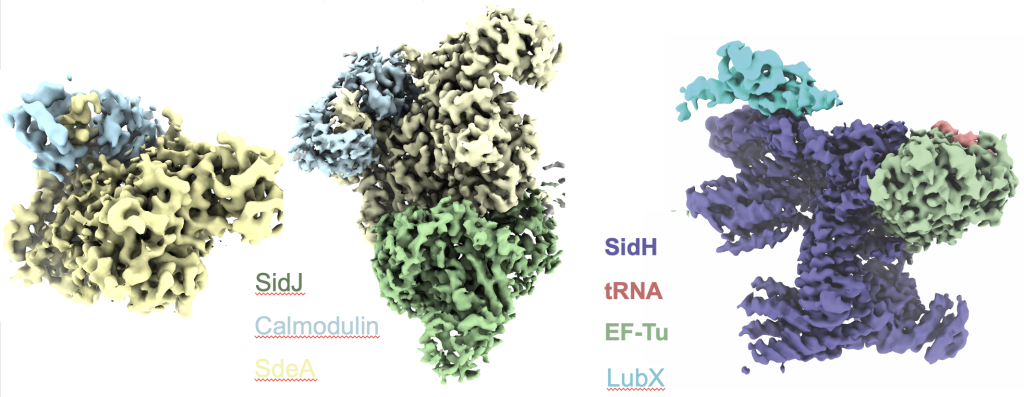
Figure: Cryo-EM structures of Legionella effector complexes. left, SidJ in complex with human Calmodulin; middle, SidJ/Calmodulin/SdeA complex; right, SidH in complex with bacterial EF-Tu, tRNA and the ubiquitin ligase LubX.
Future projects and goals:
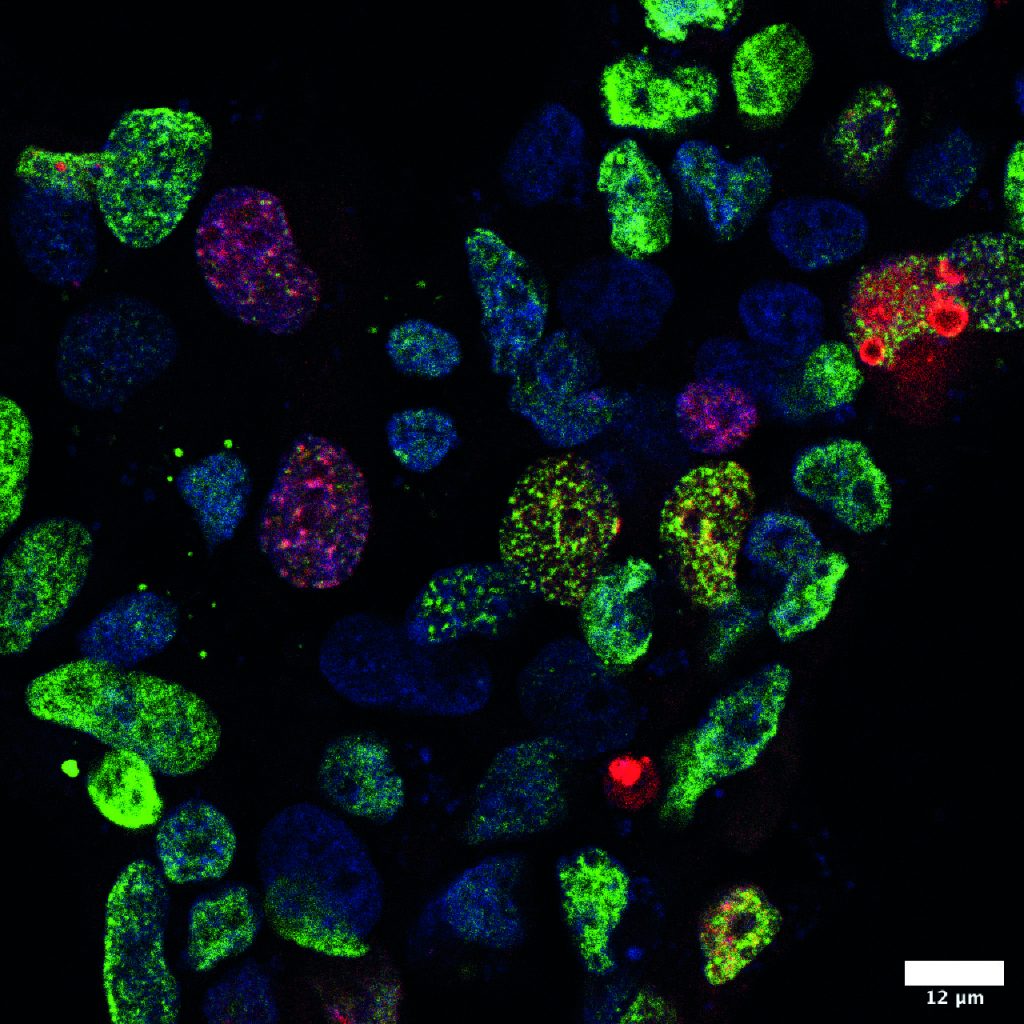
Figure: Utilizing the Ub-POD, a method we developed to identify substrates of ubiquitin ligases, we directly visualized the ubiquitination activity of RAD18 in cells in response to UV treatment. Red-Biotin; Blue-DAPI; Green-H2AX, a DNA damage marker