New findings reveal how influenza virus hijacks human cells
Scientists provide atomic resolution details of a promising drug target in influenza virus
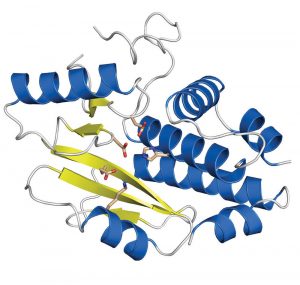
Influenza is and remains a disease to reckon with. Seasonal epidemics around the world kill several hundred thousand people every year. In the light of looming pandemics if bird flu strains develop the ability to infect humans easily, new drugs and vaccines are desperately sought. Researchers at the European Molecular Biology Laboratory (EMBL) and the joint Unit of Virus Host-Cell Interaction (UVHCI) of EMBL, the University Joseph Fourier (UJF) and the National Centre for Scientific Research (CNRS) in Grenoble, France, have now precisely defined an important drug target in influenza. In this week’s Nature they publish a high-resolution image of a crucial protein domain that allows the virus to hijack human cells and multiply in them.
When the influenza virus infects a host cell, its goal is to produce many copies of itself that go on to attack even more cells. A viral enzyme, called polymerase, is key to this process. It both copies the genetic material of the virus and steers the host cell machinery towards the synthesis of viral proteins. It does this by stealing a small tag, called a cap, from host cell RNA molecules and adding it onto its own. The cap is a short extra piece of RNA, which must be present at the beginning of all messenger RNAs (mRNAs) to direct the cell’s protein-synthesis machinery to the starting point. The viral polymerase binds to host cell mRNA via its cap, cuts the cap off and adds it to the beginning of its own mRNA – a process known as ‘cap snatching’. But exactly how the polymerase achieves this and which of the three subunits of the enzyme does what, has remained controversial.
Researchers of the groups of Rob Ruigrok at the UVHCI and Stephen Cusack at EMBL have now discovered that part of a polymerase subunit called PA is responsible for cleaving the cap off the host mRNA.
“Our results came as a big surprise, because everybody thought that the cleaving activity resides in a different part of the polymerase,” explains Rob Ruigrok, Vice-Director of the UVHCI.
“These new insights make PA a promising antiviral drug target. Inhibiting the cleaving of the cap is an efficient way to stop infection because the virus can no longer multiply. Now we know where to focus drug design efforts,” adds Stephen Cusack, Head of EMBL Grenoble and Director of the UVHCI.
The researchers produced crystals of the crucial PA domain and examined them with the powerful X-ray beams of the European Synchrotron Radiation Facility (ESRF) in Grenoble. The high-resolution image of the domain reveals the individual amino acids that constitute the active site responsible for cleaving the RNA; information that could guide the design of future antiviral drugs.
Only a few months ago, the same group of scientists had already identified another key part of the influenza polymerase; a domain in the subunit called PB2 that recognises and binds to the host cap. Taken together the two findings provide a close-to-complete picture of the cap snatching mechanism that allows the influenza virus to take control over human cells.
The research was funded by the European Commission, as part of the FP6 initiative FLUPOL, and the French Agence National de Recherche, as part of the initiative FLU INTERPOL.



