
A tRNA modifier at work
Cryo-EM reveals the structure of a piece of cellular machinery with important medical implications
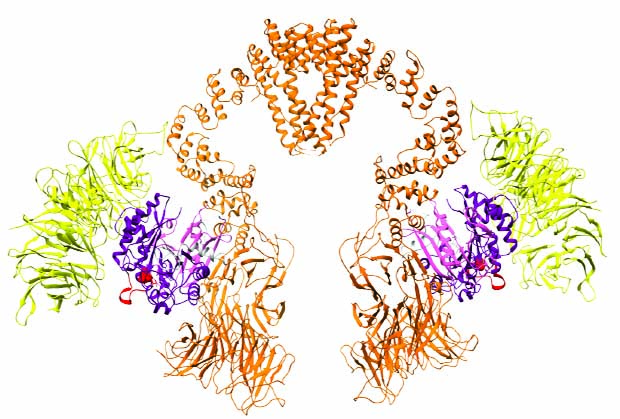
All living cells employ ribosomes and transfer RNA (tRNA) molecules to decode messenger RNA (mRNA) sequences and translate the genetic information into correctly assembled and functional proteins. It’s crucial for cells to produce these proteins at the right speed to avoid protein misfolding and aggregation within cells, which has been linked to a number of neurodegenerative diseases and cancer. The modification of tRNA molecules by specialised enzymes is one way to regulate the production speed to the correct pace.
Elongator: a large protein complex
Scientists from the Müller group at EMBL Heidelberg and the Kosinski group at EMBL Hamburg, in collaboration with researchers at the Małopolska Centre of Biotechnology (MCB) at the Jagiellonian University in Kraków, Poland, and at Martin Luther University Halle-Wittenberg, Germany, have now managed to catch a crucial component of the tRNA modification machinery directly at work. Using cryo-electron microscopy (cryo-EM), they have determined the structure of a large protein complex called Elongator, in the presence and absence of its natural substrate tRNA.
Their results, published in Science Advances, are valuable for the protein synthesis research community. They allow for a more complete understanding of one of the key mechanisms in ribosome-mediated protein synthesis, where tRNA plays a central role as an adaptor molecule – it forms the physical link between mRNA and the amino acids to be assembled into proteins.

Medical implications
“After 10 years of research on this large cellular machine, we finally start to have a clearer picture of the complicated interplay between the different Elongator subunits and the bound substrate tRNA, which permits the modification reaction,” says group leader Christoph Müller.
Mutations in any of the six subunits of Elongator are associated with the onset of human diseases, including familial dysautonomia (a disease of the nervous system), obesity, bronchial asthma, intellectual disability, ventricular hypertrophy, Rolandic epilepsy and cancer. Thus, its potential as a drug target is ripe for exploration. “Our structural work tells us how the tRNA is specifically positioned in the active site of the complex, but also allows a structure-based design of specific drug molecules that might cure Elongator-related diseases in the future,” says co-author Sebastian Glatt, a former postdoc in the Müller group, who now leads the Max Planck Research Group at MCB. Glatt is currently involved in setting up the first cryo-EM facility in Poland – an EMBL member state since February this year.
Integrative modelling
For the Kosinski group, the cryo-EM structure of Elongator marks another success. Two years ago, they published a model of Elongator built using a computational modelling method developed by Jan Kosinski. Comparing this model with the high-resolution cryo-EM structure shows a remarkable similarity between them – a clear demonstration of the power of so-called integrative modelling methods, which model structures based on data from various experiments. “We are very excited by the accuracy of the model, and are now working towards publishing our methods as easy-to-use software,” says Jan Kosinski. The group is also organising an EMBO Practical Course, to take place in November, where such integrative methods will be taught.
Register for the EMBO Practical course, ‘Practical integrative structural biology’, 3–9 November 2019
Register for the EMBL Course, ‘Cryo-Electron Microscopy and 3D Image Processing’, 23–31 August 2020


