Mapping the human brain: a global collaboration
NIH BRAIN Initiative to fund brain atlases, network coordination, and knowledge sharing to explore brain function research
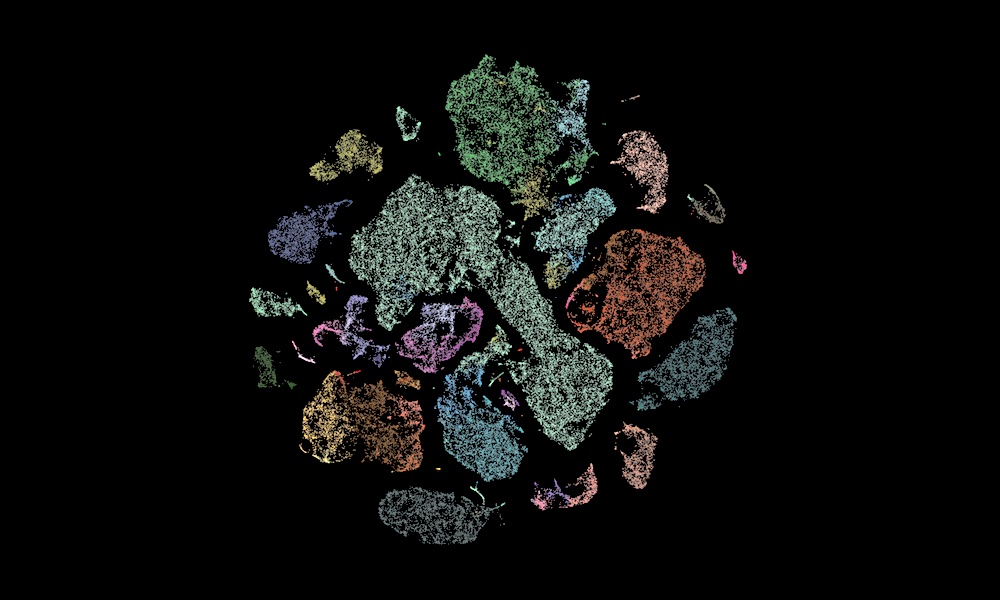
A new global collaboration will map the approximately 200 billion cells in the human brain by their type and function, and also build detailed atlases of macaque and marmoset monkey brains.
The collaboration is funded by the National Institutes of Health as part of the BRAIN Initiative® Cell Atlas Network (BICAN). This collaborative effort led by the Allen Institute also involves researchers from 17 other institutions in the US, Europe and Japan including EMBL’s European Bioinformatics Institute (EMBL-EBI).
“We are aiming to create something transformative for the field that can only be done collaboratively, by bringing in an all-star cast of experts from a variety of disciplines,” said Ed Lein, Senior Investigator at the Allen Institute for Brain Science. “This is critical work; we need to understand the human brain better if we hope to treat diseases of the brain, and specifically, we need a better understanding of brain function and structure. The cell atlases we’re building with the support of the BRAIN Initiative promise to lead to a more rapid understanding of the basis of many brain diseases.”
Data management at scale
Researchers at EMBL-EBI are involved in one of the five BICAN grants awarded to the Allen Institute. Together, the five awards total over $173 million in funding to support data generation and analysis, coordination, and administration of the network. The findings, techniques, and data from these projects will be made publicly available.
This work will bring together data from different state-of-the-art sequencing methods to create a high-resolution map of the brain. The brain is very complex and made up of many different cell types. Collecting, standardising, and storing these data at scale is therefore very challenging.
“There will be a huge amount of data coming from this project,” said David Osumi-Sutherland, Ontologies Coordinator at EMBL-EBI. “Many different groups from different institutions across the world will be generating and analysing these data. Coordinating this will require careful data management. EMBL-EBI will play three key roles: standardising the metadata that is critical to organising and integrating the diverse data generated by this project, providing tools and support for annotating the results of data analysis, and guiding the production of the final catalogue of different brain cell types, which will be openly available for the global scientific community.”
From mice to humans
These projects build on earlier NIH BRAIN Initiative-funded projects at the Allen Institute to map the cells of the entire mouse brain and parts of the human brain using single-cell transcriptomics. Understanding the cell types in commonly studied mammals like mice is key to improving translational research to ultimately benefit people suffering from brain diseases and disorders.
The primate brain atlas project will also marry cellular maps with brain function using a technique known as functional MRI that captures blood flow in the brain; active regions of the brain are associated with higher levels of blood flow. The researchers will then correlate these active regions with specific cell types using an emerging method known as spatial transcriptomics, which labels cell types in their original location in the brain. The human atlas researchers will also expand experiments that study the details of living human neurons using brain tissue donated by Seattle-area brain surgery patients.
Read the full press release on the Allen Institute website.
Find out about EMBL’s centre for epigenetics and neurobiology research.