
Breaking the bottleneck in imaging data collection
Scientists develop software tools for automated acquisition of electron microscopy data
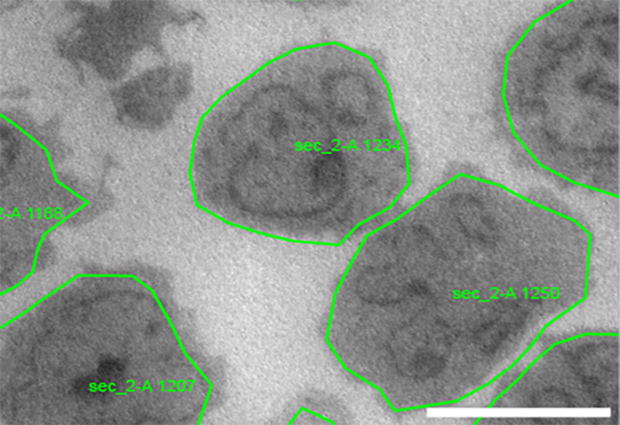
There is a growing demand for high-throughput data acquisition in structural and cell biology research. Developments in microscopy hardware and computing performance have increased the speed and quality of transmission electron microscopy (TEM). However, until now, scientists have had to manually select the features of interest to be imaged, and identify their precise coordinates: a time-consuming task that creates a bottleneck in the data acquisition procedure.
Martin Schorb, Application Engineer in EMBL’s Electron Microscopy Core Facility, and colleagues from EMBL Heidelberg and the University of Colorado, USA, have developed two software tools that allow automated acquisition of electron microscopy data. The tools, SerialEM and py-EM, are presented in a paper published in Nature Methods.
SerialEM, which was developed by scientists at the University of Colorado, is a versatile software package for controlling microscopes and cameras, to allow automated acquisition of TEM data. The py-EM module – a collection of Python functions – interfaces with SerialEM to create feedback pipelines in specimen-specific image analysis. Py-EM and the applied workflows were developed at EMBL.
Using these tools, scientists no longer need to define discrete positions on an object for data acquisition. Instead, they define whole target objects, such as cells or distinct tissue features, and the tools create automated feedback TEM workflows for these objects.
The new software tools make it possible to image every entity of a specimen in a controlled way, and this opens up new possibilities for TEM applications. For example, the automated approach enables systematic screening for phenotypes within cell populations or across different conditions. It also significantly increases throughput in structural analyses of molecular complexes using cryo-electron microscopy.


