
Read the latest Issue
Scientists at EMBL Hamburg helped to reveal how the herpes simplex virus hijacks human proteins
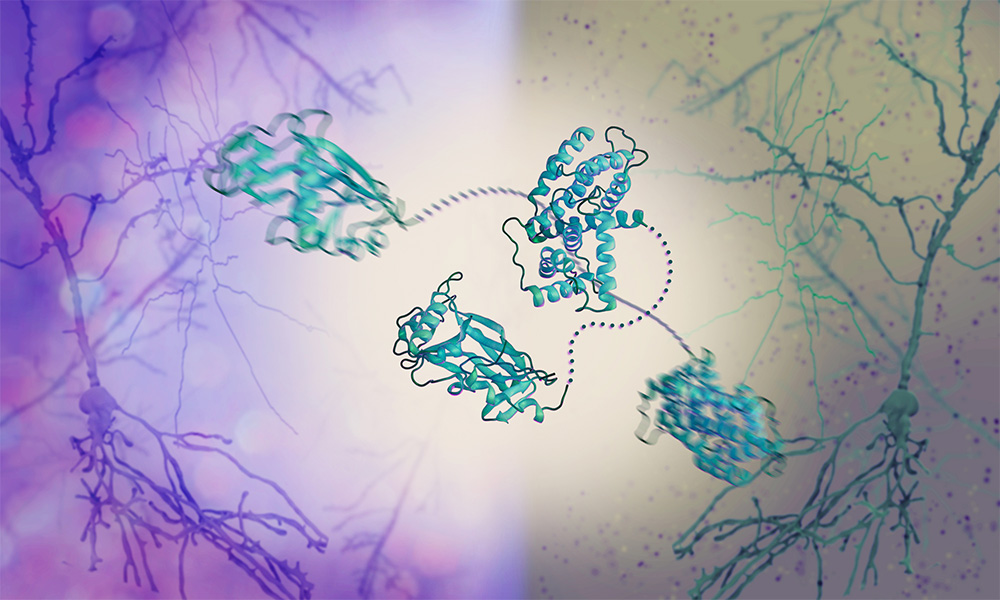
The Graham and Crump groups at the University of Cambridge and the Svergun Group at EMBL Hamburg have discovered a mechanism by which the herpes simplex virus takes control of the molecular machinery of human cells. Their work reveals how a dedicated viral protein hijacks key host proteins, forcing the host cell to produce and release copies of the virus.
Have you ever experienced a cold sore on your lip? If so, you’re not alone. The World Health Organization estimates that worldwide, 67% of people under the age of 50 are infected with herpes simplex virus (HSV-1), which is well known for causing cold sores. It can also cause genital herpes, blindness, and in rare cases, brain infection and death. Some studies suggest that HSV-1 infection may also increase the long-term risk of dementia.
Once infected, a person carries the virus with them for life. While some of the symptoms can be treated, there is currently no cure and no effective vaccine. The virus remains dormant in our nerve cells, only to reappear and cause renewed symptoms under certain conditions, such as stress.
Like other viruses, HSV-1 multiplies by hijacking the molecular machinery of infected human cells. The infected cell starts to produce copies of the virus, which then use the cell’s secretory system to exit the host cell and infect other cells. Disrupting this cycle at any stage – cell entry, virus production, or egress from the cell – could halt the spread of the virus and stop the infection.
One of the steps required for virus replication is the production of viral proteins. To ensure all parts of the molecular machinery work in synchrony, cells have dedicated ‘production managers’ – proteins that switch other proteins on and off when needed. Some of these proteins, called kinases, attach a small molecular ‘tag’ to the protein they control. Others, called phosphatases, do the opposite – they remove the tag. To reprogram a cell’s metabolism, viruses often hijack these production managers. While many studies have looked at how viruses hijack kinases, very little is known about their influence on phosphatases.
To fill this gap, the Graham and Crump groups at the University of Cambridge and the Svergun Group at EMBL Hamburg studied the effect of HSV-1 on phosphatases. The scientists revealed a new mechanism by which HSV-1 hijacks a human phosphatase. They investigated a viral protein called pUL21, which is critical for the spread of the virus, and found that it binds the phosphatase and other human proteins involved in protein secretion.
How can pUL21 be so versatile and hijack not just one but several different proteins? To answer this question, the Svergun Group used a structural biology technique called small-angle X-ray scattering (SAXS), which allows scientists to study the dynamic movements of proteins. SAXS revealed that pUL21 comes in the shape of a flexible string with rigid components at each end, which bind other proteins. It ‘chains’ the human phosphatase 1 bound at one end to another protein at the other end, just like handcuffs. By bringing the two proteins close together, pUL21 forces the phosphatase to steer the activity of the other protein, which in turn supports the production and release of viral copies from the cell.
“It’s fascinating that pUL21 can bind human proteins of different shapes and sizes. It’s because it’s so flexible,” said Dr Cy Jeffries, a senior scientist in the Svergun Group. “A protein that adapts its shape so easily can serve multiple purposes. That explains how the virus can interfere with so many cellular processes despite having only a few proteins of its own.”
“We saw that pUL21’s flexibility plays a major role in viral replication. The SAXS expertise of our long-established collaborators at EMBL Hamburg helped us uncover how this happens,” said Dr Stephen Graham. “Understanding how the viral protein behaves and interacts with human proteins may help design new drugs and vaccines against herpes.”
Infection biology, which is one of EMBL Hamburg’s areas of focus, is at the heart of the forthcoming EMBL Programme, Molecules to Ecosystems 2022–2026. As part of the programme, EMBL will take an interdisciplinary approach to understanding the molecular basis of life in the context of its environment. This will provide the fundamental science needed to support translational advances in human and planetary health.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive