Charting the landscape of cytoskeletal diversity in microbial eukaryotes
Cell 31 October 2025
10.1016/j.cell.2025.09.027
EMBL researchers have been using a powerful technique called expansion microscopy to peek deeper inside living organisms and understand how they function
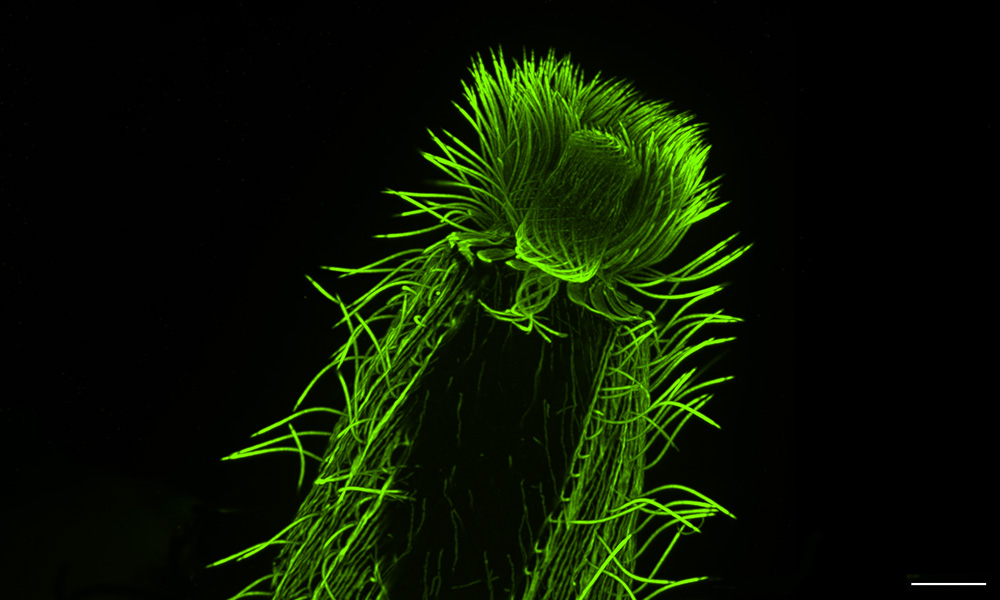
One day in 2020, in the middle of the COVID-19 pandemic, EMBL Group Leader Gautam Dey received a Zoom call from his collaborator and fellow cell biologist Omaya Dudin. Dudin, who led a research group at École Polytechnique Fédérale de Lausanne (EPFL), Switzerland, at the time, had just succeeded in optimising a relatively new technique to observe the cellular architecture of a marine microorganism.
Dudin had been working with this particular group of marine microbes – Ichthyosporea – for over six years. However, visualising its inner structures had been a challenge. Unlike for more commonly studied organisms, there were no genetic tools to make certain structures light up under the microscope. Nor did antibody-staining, the other main technique in the biologist’s toolkit, work well in this organism, as its cell wall prevented antibodies from penetrating inside the cell.
However, the new technique – expansion microscopy – had somehow made this cell wall permeable, letting antibodies enter, and the organism’s inner structures could now be clearly visualised and studied.
“Hey, we should do this with all microbial eukaryotes,” Dudin said.
Dey agreed, and thus was born a collaboration that, three years later, has succeeded in generating almost encyclopedic knowledge of hundreds of species and is on its path to creating a planetary atlas of plankton.
Microbial eukaryotes, also known as protists, are tiny organisms, usually single-celled, which have membrane-bound organelles, such as a nucleus to contain their genetic material. As the scientists found, expansion microscopy can be instrumental in yielding new biological insights into evolution and cellular function in this highly diverse group.
Developed 10 years ago by the group of Edward Boyden at MIT, and further optimised into ultrastructure expansion microscopy (U-ExM) for exploring sub-cellular ultrastructure by Paul Guichard and Virginie Hamel at the University of Geneva, expansion microscopy allows scientists to easily and inexpensively visualise cellular structures that are highly challenging to see with traditional microscopy techniques.
Dey and Dudin are not alone among EMBL scientists and collaborators in taking advantage of expansion microscopy. In addition to the Dey Group, researchers from the Banterle, Saka, and Vincent groups at EMBL Heidelberg, the Sharpe Group as well as the Mesoscopic Imaging Facility at EMBL Barcelona, and the Light Imaging Facility at EMBL Rome, are adopting expansion microscopy protocols to help answer pressing questions in cellular and developmental biology.
Expansion microscopy, as the name suggests, works by physically ‘expanding’ biological samples. These samples – which can contain single-celled organisms, cells, or tissues – are first embedded in a clear gel. Then, the gel is allowed to expand by absorbing water. Marvellously, many of the cell’s internal structures remain intact during the process and expand more or less proportionally, allowing scientists to ‘magnify’ the sample four or even 16 times without needing to resort to lenses.
“It’s a technique that, when you think about it, sounds as if it shouldn’t work,” said Dey. “But it does, spectacularly so, and it opens up a whole new world of fundamental biological questions we can now ask quickly and at scale.”
When combined with regular light microscopy methods, expansion microscopy allows scientists to bypass the standard wavelength barriers which limit how small a structure can be resolved using light microscopy.
Take centrioles, for example. These barrel-shaped organelles play an important role in cell division, help anchor cilia and flagella – hairlike protrusions that help cells move – and nucleate the network of filaments that form the cell’s inner skeleton.
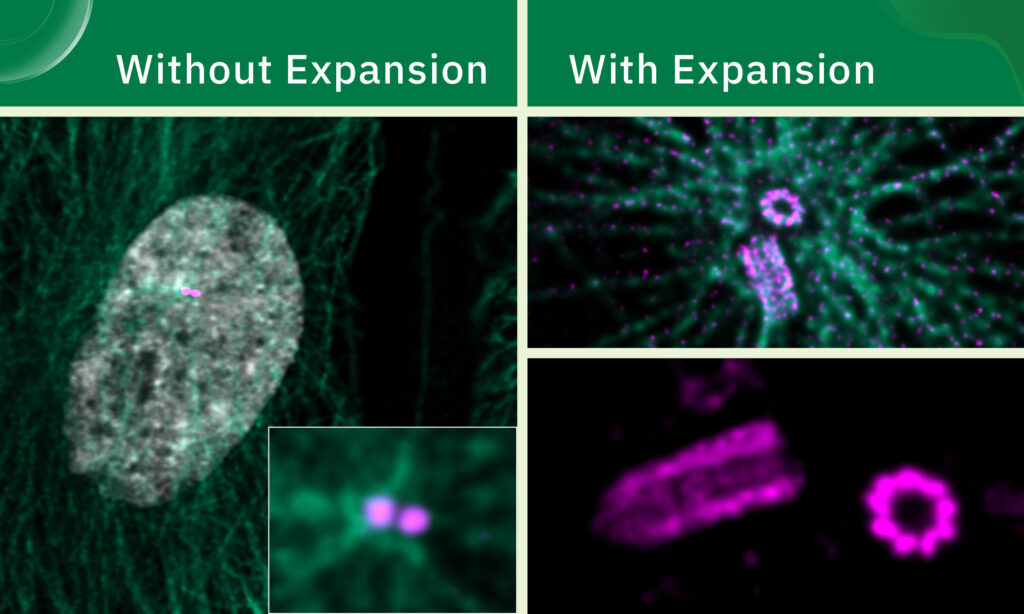
Viewed under a conventional confocal microscope, centrioles appear as tiny, formless dots. However, as EMBL Group Leader Niccolo Banterle learned during his postdoc, expansion microscopy increases the resolution of light microscopy to the level where it is possible to see the centrioles’ beautifully symmetric fine structure.
Using a combination of ultrastructural expansion microscopy and super-resolution imaging techniques such as stimulated emission depletion (STED) microscopy, Banterle’s team is investigating how factors such as centriole length and the nature of proteins that link them to each other affect cellular function.

For Dey and Dudin, however, it all comes back to microbial eukaryotes. One particular group they are interested in is plankton – tiny organisms found in the Earth’s oceans that form an irreplaceable part of the oceanic food chain and generate most of the planet’s oxygen. Their diversity already spans tens of thousands of species, and that’s only among those described so far, with many more waiting to be discovered.
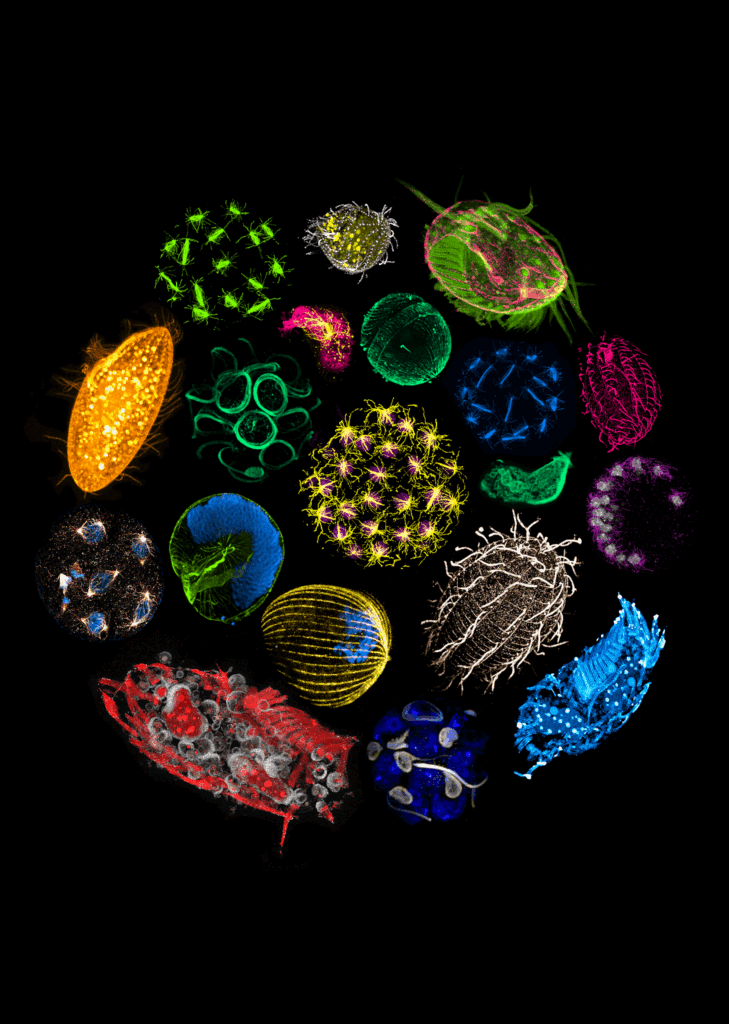
Until recently, genomic sequencing was the strongest tool in researchers’ arsenal to study planktonic diversity. Available imaging methods were either high-resolution or high-throughput, but rarely both. Many of these organisms could also not be cultured under laboratory conditions and had no genetic tools available.
“Thus, the fields of cell biology and protistology diverged over the last 40 or so years, with one focusing on increasingly high-resolution microscopy methods, while the other specialised in genomics,” Dudin explained.
For the collaborators, the first opportunity to bridge this gap came during the EMBL-led Traversing European Coastlines (TREC) expedition. One of the expedition’s first ‘superstops’ was at Roscoff, France, where the Station Biologique hosts one of the most complete culture collections of marine microorganisms in Europe. The team asked Ian Probert, the facility’s manager, how many samples they might receive to run an expansion microscopy pilot, expecting about 20 species. Instead, the facility welcomed them with more than 200 species ready to be processed for expansion microscopy.
“We spent three days and nights just fixing those samples. This was a treasure trove we could not let go of,” said Felix Mikus, who completed his PhD from the Dey Group and is now a postdoc in Dudin’s current lab at the University of Geneva.
Using these samples, as well as a second culture collection from Bilbao, Spain, the researchers went on to perform one of the most extensive investigations to date of the diversity of the cytoskeleton – the filamentous network that forms the underlying structure of eukaryotic cells. In particular, researchers looked at microtubules – hollow, tubular filaments that help the cell not only maintain its structure, but also divide and move. They also studied centrins, a family of proteins found in the structures that organise microtubules inside cells.

Recently published in Cell, this study was the first step in PlanExM, a TREC plug-in project that aims to reveal the world of planktonic ultrastructural diversity with expansion microscopy.
“We were able to map features of microtubule and centrin organisation across many different eukaryotic groups,” said Hiral Shah, co-first author of the study and EIPOD Postdoctoral Fellow in EMBL’s Dey and Schwab groups, as well as in the Dudin Group in the University of Geneva. “The scale of the study, with many species characterised in each group, opens up the possibility to make evolutionary predictions. For instance, dinoflagellates, one of the most diverse groups found in oceans across the planet, are well-represented in our study. We were able to map the presence of tubulin and centrin structures associated with the cell cortex or the flagella in these species.”
According to the researchers, this analysis not only helps us understand the fundamental principles that underlie the organisation of a eukaryotic cell, it also offers clues to the evolutionary history of the cytoskeleton architecture in these species. Moreover, it demonstrates the potential of expansion microscopy as a powerful tool to study complex samples, including those collected directly from marine ecosystems.
“U-ExM is transforming how we explore protist ultrastructure,” said Armando Rubio Ramos, co-first author of the study and postdoc at Hamel & Guichard’s research group at the University of Geneva. “By combining this technique with high-throughput imaging and comparative analyses, we can begin to decode how cellular architecture has diversified across evolution. It’s a bridge between molecular data and the physical organisation of life at the microscopic scale.”
With guidance from Thomas Richards from Oxford University, Dey and Dudin were also successful in obtaining a prestigious Moore Foundation Grant worth CHF 2 million to continue this project. “The next step is to selectively look deeper into certain species within this broad collection to answer specific questions, such as understanding how mitosis and multicellularity evolved and the phenotypic diversity that underlie major evolutionary transitions,” Dudin said.
Expansion microscopy can also help scientists better investigate and understand interactions between organisms ‘in the wild’, i.e. in their natural environment. Flora Vincent, another EMBL Group Leader, is also an integral part of TREC, trying to further her studies on symbioses among marine microbial species, especially those belonging to a group called diatoms.
“Diatoms are small microscopic organisms that live in the ocean and play a huge role in our ecosystem, because they produce very large amounts of oxygen every year that are critical for life,” Vincent said. “They also form the basis of the food chain, because grazers feed on them. So understanding how and why diatoms can actually multiply so fast, and with whom they interact, can help us understand what sustains life in the ocean.”
Serena Flori, a postdoc in the Vincent lab, started using expansion microscopy to study diatoms about two years ago with support from collaborators in the Dey Group. “Diatoms are unique in having a special type of cell wall, made out of silica, which makes it very difficult to study them using antibodies,” she explained. To her delight, by rapidly freezing diatom samples and then imaging them using ultrastructural expansion microscopy (cryo-ExM), the team could overcome this ‘silica barrier’. The new approach allowed Flori to study the cellular structures she was interested in without resorting to alternative, more time- and cost-intensive electron microscopy methods.
The process also worked for environmental samples collected from the field during the TREC expedition, and the team could study diverse diatom species spanning over 80 million years of evolutionary time, gaining new insights into variations in the cytoskeleton and photosynthetic machinery, as well as into the symbiosis within diatom species. The team recently published their results in the journal Current Biology.
Beyond PlanExM, Dey Group members have also been using expansion microscopy to answer fundamental questions about evolution in eukaryotes. Hiral Shah expertly used the technique to show how different modes of cell division in animals and fungi might have evolved to support diverse life cycles. Once more through a collaboration between the Dey and Dudin Groups, the team studied two different species of Ichthyosporea, which are close relatives of both animals and fungi, using ultrastructural ExM to tease out molecular differences in how they performed cell division.
“The first time we saw an expanded Ichthyosporean nucleus, we knew this technique would change the way we study the cell biology of non-model organisms,” said Shah, who brought back the expansion microscopy technique to the Dey Group after a stint at the Dudin lab.
Mikus, as part of his PhD work in the Dey Group, helped standardise the ExM technique for budding and fission yeast – two important model organisms for microbiologists. He went on to use this to explore subcellular organisation in both types of yeast and show its link to cell division.
Not everything expanded by ExM needs to be minute, however. Marie Jacobovitz, postdoctoral fellow in the Dey Group, is using expansion microscopy to interrogate a fascinating microorganism – Physarum polycephalum – a slime mould that can grow to such proportions that it is easily visible with the naked eye. Jacobovitz hopes ExM provides a clearer view of the Physarum environment, so she can better understand its impact on regulating its cell cycle. She also hopes to combine ExM with other imaging modalities, such as electron microscopy, for more detailed structural understanding of the molecular processes involved.

Scientists have also begun to explore the potential of expansion microscopy in imaging larger tissues and structures, going beyond unicellular organisms and cells grown in the lab. Laura Campamà, an intern in EMBL Barcelona’s Mesoscopic Imaging Facility (MIF), spent a couple of months standardising the technique to work on mouse embryos to study cellular organisation during tissue growth. In this, she was guided by Montserrat Coll Lladó, Mesoscopic Imaging Scientist.
Among those making use of ExM approaches on mouse embryos is Heura Cardona Blaya, Laboratory Manager from the Sharpe Group in EMBL Barcelona. The Sharpe Group unites experimental and computational approaches in studying a critical process – the formation of limbs and digit patterning during mammalian development. Cardona is using and optimising expansion microscopy to study how cells orient themselves across the limb during development. Without ExM, one would need to physically section the limbs and image them at high resolution. However, in this process, one loses spatial 3D information and the cell environment.
“A big advantage of this technique is that you can image at high resolution small features from cells, even in thick and packed tissues such as the mouse limb,” said Cardona, “Moreover, this makes it possible to study not only microscopic but also bigger organisms or tissues such as embryos or tumours. Our hope is that the data collected using this method will eventually help us reconstruct cell orientation during limb development in 3D.”
Researchers are also trying to combine ExM with other advanced spatial imaging techniques, bringing the best of multiple worlds together to understand biological processes at a molecular level.
Sinem Saka’s group at EMBL Heidelberg specialises in using labelling technologies that combine DNA nanotechnology with advanced imaging modalities. To this end, Kristina Jevdokimenko, an ARISE fellow in the Saka Group, is working on combining expansion microscopy with a technique called Immuno-SABER.
This method allows scientists to tag multiple types of molecules inside cells and tissues with barcodes made out of DNA. These barcodes can then be read out with fluorescence imaging either over cycles of detection or with spectral multiplexing. In the latter case, which is more applicable to samples with large volumes, signals from different fluorescent molecules can be ‘unmixed’ through advanced computational methods to allow scientists to detect up to 15 different types of molecules in one imaging round. When applied in combination with expansion microscopy, this approach allows simultaneous visualisation of many different subcellular structures at once with molecular resolution and high efficiency.
Expansion microscopy is a relatively young technique, only turning 10 years old this year, but it is spreading rapidly, aided by the researchers who have been using it regularly. Each of the above research teams routinely receives requests for collaborations and training, and they have made it their mission to try and accommodate as many of those requests as possible. In 2024, the researchers co-organised an EMBL | EMBO practical workshop to teach this technique to scientists from across Europe, who were planning to apply the method to their own unique research problems. The workshop will be offered again at EMBL Heidelberg in 2026, co-organised by Dey and Banterle, alongside Paul Guichard and Virginie Hamel from the University of Geneva.
“Our adventures with expansion microscopy are only beginning,” said Dey. “This is perhaps the first high-resolution microscopy technique that has the potential to match the scale and ambition of large biodiversity genomics projects, enabling us in the near future to associate new multiomics data with cellular physiology at scale across the tree of life.”
Cell 31 October 2025
10.1016/j.cell.2025.09.027
Current Biology 31 October 2025
10.1016/j.cub.2025.10.024