
Niccolò Banterle
Group Leader
ORCID: 0000-0002-1607-0324
EditUnderstanding cross-scale principles of cellular organisation

Group Leader
ORCID: 0000-0002-1607-0324
EditLiving systems maintain order at different spatio-temporal scales, ranging from individual molecules up to tissues and ultimately multicellular organisms. Even within single cells, there are several scales at which organisation arises: from protein folding at the 10 nm scale to macromolecular assemblies at the 100 nm scale, up to cellular compartments and cytoskeletal organisation at the 1–10 mm scale. In recent years, scientists’ understanding of the biophysical laws describing the emergence of well-defined structures at all these scales has seen major advances. Yet, for healthy cell physiology, coordination between processes at different scales is required. We investigate how these scales influence each other, from the molecular to the physiological level.
Improper assembly of macromolecular complexes can drastically affect cellular functions. How does the ultrastructure of complexes dictate these functions? We tackle this key biological question by focusing on centrioles. Centrioles are microtubule-based macromolecular complexes with defined symmetry, dimensions, polarity, and chirality. By recruiting the pericentriolar material, centrioles form the centrosome: the major microtubule-organising centre of animal cells. The centrosome is crucial for organising the cytoskeleton, and therefore influences many cellular functions, such as cell division, polarity, motility, and immune synapse formation. Changing centriole ultrastructure results in altered cellular physiology, and as such is an ideal model for connecting the 100 nm ultrastructure, the 10 mm cytoskeletal organisation, and ultimately the cellular functions. Combining the latest developments in super-resolution imaging and high-speed atomic force microscopy opens the possibility of bridging these scales. Applying these and other biophysical methods together with live-cell imaging, we will unravel changes in structure and dynamics across scales and integrate them in a multiscale model to connect centriole ultrastructure with cell physiology.
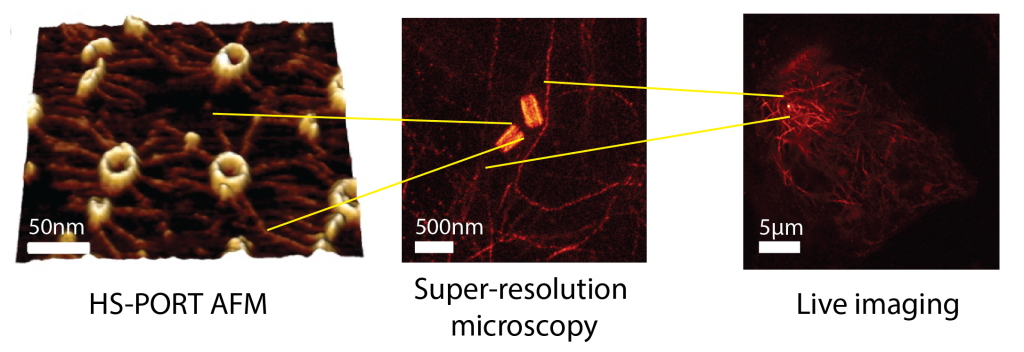
We will initially focus on perturbing three known centriole ultrastructural variables and will acquire multiscale in vitro and in vivo datasets to unravel the fundamental mechanisms explaining: