
Read the latest Issue
EMBL scientists reveal detailed insights into self-splicing introns
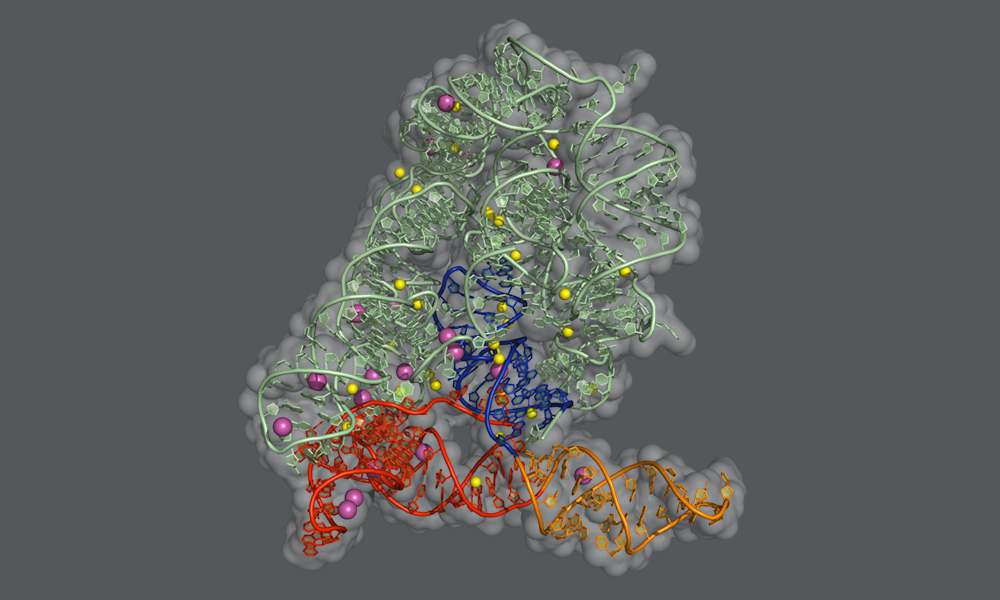
When the Human Genome Project was completed, many scientists were surprised to discover how few genes we have. Humans have about as many genes for making proteins as mice or roundworms – around 20 000 – and significantly fewer than crops like rice or corn. One way of achieving more biological complexity with a comparatively small number of genes, it turns out, is through a process called splicing. Researchers in the Marcia group at EMBL Grenoble and collaborators in the De Vivo lab at the Istituto Italiano di Tecnologia (Italian Institute of Technology, IIT) in Genoa have now obtained some of the most detailed ever snapshots of the splicing process in systems known as group II self-splicing introns. Their findings are published in Nature Communications.
Splicing is a crucial step in using the genetic information stored in DNA to make proteins, which carry out the majority of biological functions. To make genetic information usable, DNA has to be converted, or transcribed, into messenger RNA (mRNA) – a molecule that acts as a transient carrier of information. In animals, most genes and their mRNA transcripts contain long sequences of genetic information that are not used directly to make part of a protein. These segments, which are called introns, have to be cut from their mRNA strand before it can be used to produce a functional protein. By cutting different combinations of introns from an mRNA, cells can also produce different protein variants from a single gene. This allows organisms to increase the number of proteins they can make, without having to carry a separate gene for each protein, keeping the genome compact.
The system is comparable to an elaborate secret language: the actual message is interrupted multiple times by a large number of filler words. Some of these words contain instructions about which parts of the message need to be removed and how the remaining ones have to be reconnected to obtain a meaningful sentence – a spliced mRNA that can produce a functional protein. It’s essential to precisely cut out the filler words (introns) and paste together the remaining pieces (called exons). In most cases, any mistakes in splicing will produce a sentence that is incomplete or doesn’t make sense – the end result is a protein that cannot carry out its intended function.
In humans and many other organisms, the splicing process is carried out by a molecular machine called the spliceosome, which is a complex assembly of RNA molecules supported by proteins. In bacteria and other microbes, as well as in subcellular structures in plants and fungi, the introns themselves fold into a particular shape that enables them to cut themselves out from their precursor mRNA. “These self-splicing introns, called group II introns, are the evolutionary ancestors of the RNA units of the spliceosome, and the two machineries use the very same chemical reaction to perform splicing,” explains Jacopo Manigrasso, PhD student in the De Vivo lab and one of the two lead authors of the study. “By understanding the self-splicing mechanism of group II introns, we can better understand how splicing works in humans as well.”
In this study, the Marcia group at EMBL Grenoble and the De Vivo lab at IIT used X-ray protein crystallography and computer simulations to figure out the atomic details of the group II intron splicing reaction. “Each splicing event consists of two major steps: a cut at one end of an intron, followed by a cut at the other end that releases the intron and simultaneously reconnects the two flanking exons of the mRNA,” explains Isabel Chillón, Postdoctoral Fellow in the Marcia group, who shares lead authorship of the study. “In between these steps, the intron needs to be slightly rearranged – but the precise mechanism of this rearrangement has not been known until now.”
The scientists have now found that the first step of the splicing reaction unleashes a positive charge within the intron. This leads to the temporary detachment of a potassium ion, which assists splicing and stabilises the intron structure, and causes a flip of the intron in the proximity of the cutting site. This transient rearrangement is crucial to establish a suitable configuration for the intron to perform the second step of the splicing reaction. When the scientists prevented the positive charge transfer in their experiments, they found that the sequence of rearrangements does not take place, and the second ‘cut and paste’ step of the splicing reaction happens at an impractically slow rate.
Apart from giving us a better understanding of the splicing reaction, the findings of this study also have implications for biotechnology and drug development. Unlike introns that are spliced out by the spliceosome and degraded, group II introns remain active molecular machines after they have been released from their mRNA. They can cut other RNA or DNA molecules and insert themselves into them, in a reversal of the splicing reaction. This feature can be used by scientists for gene editing applications, or in the development of gene-specific therapeutics. The detailed structural insights will also help scientists to develop small molecules, called splicing inhibitors, which selectively block group II intron splicing without affecting the spliceosome. Since self-splicing introns are essential for the survival of certain pathogens, such inhibitors could in the future be used to treat infectious diseases, expanding our toolkit of antibiotics.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive