
The shape of things to come
As structural biologists tackle ever larger and more complex proteins, the databanks that store the information they uncover have to find new ways of handling and distributing data.
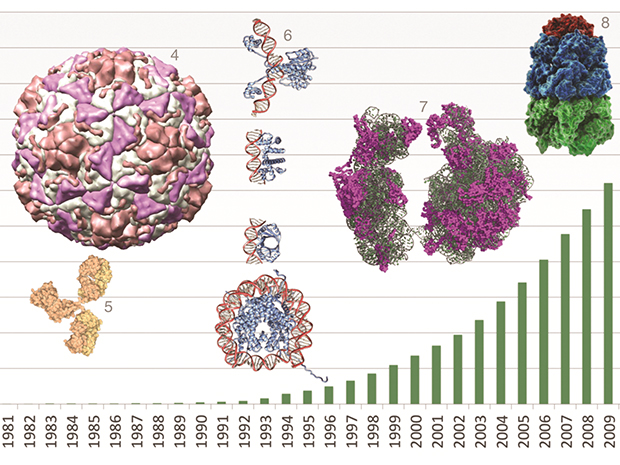
The Protein Data Bank in Europe (PDBe), run by EMBL-EBI, is in the process of launching a new website and data-handling innovations to enhance the amount of information researchers can glean from protein structure data. A timely step, as the number of protein structures in the database reached 100,000 in May this year (Another structural biology milestone was reached in Grenoble just months earlier, with the 10 000th biological structure determined at the European Synchrotron Radiation Facility, ESRF).
One innovation is the rollout of a common data deposition system that allows researchers to upload information about very large proteins in one go, using a file format called mmCIF. Previous systems could only handle smaller datasets, meaning that information about large, complex proteins was fractured into several sections. The new system will also allow researchers to integrate structural information from a range of methods, such as X-ray crystallography and electron microscopy. “Structural biologists are basically using whatever is at their disposal to solve a structure,” says Gary Battle, Outreach Coordinator for the PDBe. “The challenge for us is to cope with all of these hybrid methods.”

Another key aim is to integrate structural data with other information about a protein, such as its amino acid sequence, says Sameer Velankar, a Team Leader at the PDBe. “The main aim is to bring structure into its biological context so that you can understand more about the biological relevance and function of that structure in that system,” he says.
The main aim is to bring structure into its biological context so that you can understand more about the biological relevance and function of that structure in that system
The team has also developed a system that allows users to assess, or validate, the accuracy of the information about a protein. This system will form part of the new website, which will be launched in beta version by the end of 2014.
Listen in as Velankar and Battle from PDBe look forward to the “Google maps” of protein databases:


