Design of expression construct
To design a successful protein expression construct, it’s imperative to collect as much information as possible about your protein of interest. Some key points you need to consider are the following:
- Is it a single- or multi-domain protein? If it’s a multi-domain protein, do you want to express full-length protein or would a specific domain be sufficient for your planned down-stream applications?
- What is the native localization of the protein in the cell? Is it a soluble protein or a membrane protein? Does the native sequence contain a signal sequence?
- Does the protein require certain co-factors for stability and/or functionality? If yes, it might be necessary to add these to the culturing medium and/or the purification buffers.
- Does the protein contain a large number of disulfide bonds?
- Does the protein need interaction partners for stability? If the protein isn’t stable on itself, it might be necessary to co-express with interaction partners.
- Does the protein contain post-translational modifications? If yes, are these necessary for the folding and/or the functionality of the protein?
If there is not much information about your protein available in literature or on Uniprot, there are many bio-informatics tools that can help you predict several protein characteristics and thereby guide the construct design. Multiple sequence alignments are very helpful to define conserved parts and can help define domain boundaries, together with domain and structural predictions.
Useful bio-informatics tools
Expasy: Swiss Bioinformatics Resource Portal. Here you can find a large variety of tools and databases that are very helpful when working with proteins (for example sequence analysis tools, topology predictions, post-translational modifications predictions, 2D and 3D structural predictions, domain predictions, …)
Clustal Omega: Multiple Sequence Alignment program
SignalP: predicts the presence of signal sequences and their cleavage sites
The next step in the design of the expression construct is the choice of your expression vector. For commonly used host organisms, large collections of expression vectors are available commercially or in non-profit plasmid repositories such as Addgene and Gene Corner. We also have a collection of expression vectors that were created at EMBL, which we share freely with the academic community via a Material Transfer Agreement.
Another important point in the construct design is the decision of which protein tags to include in your construct. In many cases, an affinity tag to facilitate the purification (and/or detection) will be added. Small affinity tags (e.g. His6, StrepII, twinStrepII, Flag, Myc, HA, SPOT, …) are usually added to the N- or C-terminus of the protein, although in rare cases they can also be placed inside internal loops. Solubility-enhancing tags (e.g. SUMO, Trx, NusA, DsbA, DsbC, …) are generally placed at the N-terminus of a protein. Some protein tags such as GST and MBP can fulfil both functions at once and act both as a solubility-enhancing tag and affinity tag that can be used later on during the protein purification. Fluorescent tags (e.g. eGFP, mCherry, YFP, CFP, …) can be added to the N- or C-terminus of the protein and used for imaging purposes or studying interactions via biophysical techniques based on fluorescence. Modular tags such as HALO, SNAP and CLIP allow the attachment of different chemical functionalities and can be used to couple the protein covalently to a fluorescent dye, an affinity handle or a solid surface.
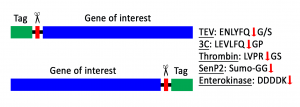
Affinity tags or solubility-enhancing tags can be removed during the protein purification when a specific protease cleavage site is included between the tag and the protein of interest.
References
Los G.V., Encell L.P., McDougall M.G., Hartzell D.D., Karassina N., Zimprich C., Wood M.G., Learish R., Ohana R.F., Urh M., Simpson D., Mendez J., Zimmerman K., Otto P., Vidugiris G., Zhu J., Darzins A., Klaubert D.H., Bulleit R.F. and Wood K.V. (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 3(6):373-82
Gautier A., Juillerat A., Heinis C., Corrêa I.R. Jr, Kindermann M., Beaufils F. and Johnsson K. (2008) An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 15(2):128-36
Saccardo P., Corchero J.L. and Ferrer-Miralles N. (2016) Tools to cope with difficult-to-express proteins. Appl Microbiol Biotechnol. 100:4347–4355
Bell M.R., Engleka M.J., Malik A. and Strickler J.E. (2013) To fuse or not to fuse: What is your purpose? Protein Science 22:1466-1477