The Gateway cloning technology is based on the site-specific recombination system used by phage λ to integrate its DNA in the E. coli chromosome. Both organisms have specific recombination sites, which are called attP in the phage λ and attB in E. coli. The integration process (lysogeny) is catalyzed by 2 enzymes: the phage λ encoded protein Int (Integrase) and the E. coli protein IHF (Integration Host Factor). Upon integration, the recombination between attB (25 nt) and attP (243 nt) sites generate attL (100 nt) and attR (168 nt) sites that flank the integrated phage λ DNA.
The process is reversible and the excision is again catalyzed by Int and IHF in combination with the phage λ protein Xis (excisionase). The attL and attR sites surrounding the inserted phage DNA recombine site-specifically during the excision event to reform the attP site in phage λ and the attB site in the E. coli chromosome.
The Gateway cloning reactions are in vitro versions of the integration and excision reactions. To make the reactions directional, two slightly different and specific sites were developed (called att1 and att2) for each recombination site. These sites react very specifically with each other. For example, in the BP reaction, attB1 only reacts with attP1, resulting in attL1 and attR1, and attB2 only reacts with attP2, leading to attL2 and attR2. The reverse reaction (LR Reaction) shows the same specificity.
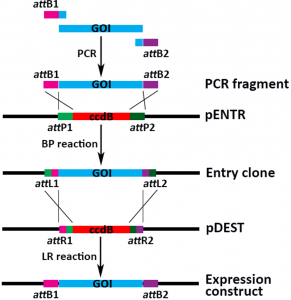
Gateway cloning is generally a 2-step process: in the first step an entry clone is generated using the BP reaction and in a second step the gene of interest is moved to a destination vector using the LR reaction.
Once you have generated a PCR fragment containing your gene of interest flanked by attB1 and attB2 sites, you can mix this with a Gateway entry vector and the BP Clonase® enzymes, leading to an entry clone in which the gene of interest is flanked by the attL1 and attL2 sites. The Gateway destination vectors contain a ccdB cassette (the encoded protein is toxic for most E. coli strains) flanked by the attR1 and attR2 sites. Upon mixing the entry clone and the destination vector with the LR Clonase enzymes®, you’ll generate an expression construct in which your gene of interest is flanked by the attB1 and attB2 sites. Negative selection by the toxic ccdB protein and different antibiotic resistances between the entry clone and the destination vector will lead to a very high level of positive clones after transformation into an E. coli strain.
The multitude of destination vectors that are available provides a lot of flexibility to generate many different expression constructs in parallel. However, there can be some disadvantages as well to Gateway cloning, such as the long N-terminal overhangs that are created with some destination vectors. If you desire to remove these overhangs fully from the mature protein, it might be advisable to introduce an extra protease cleavage site behind these overhangs (e.g. by including this into your PCR primers).
More information about Gateway cloning and available Gateway vectors, can be found on the Thermo Fischer Scientific website.