Highly accurate protein structure prediction for the human proteome.
Nature 22 07 2021
10.1038/s41586-021-03819-2
Disruptive scientific breakthroughs raise more questions than they answer. They open new research avenues and can inspire entirely new fields of study. Just as the Human Genome moment marked the beginning of a revolution in genomics, so too AlphaFold might usher in a new era in biology.
By Prof. Dame Janet Thornton, Director Emeritus of EMBL-EBI
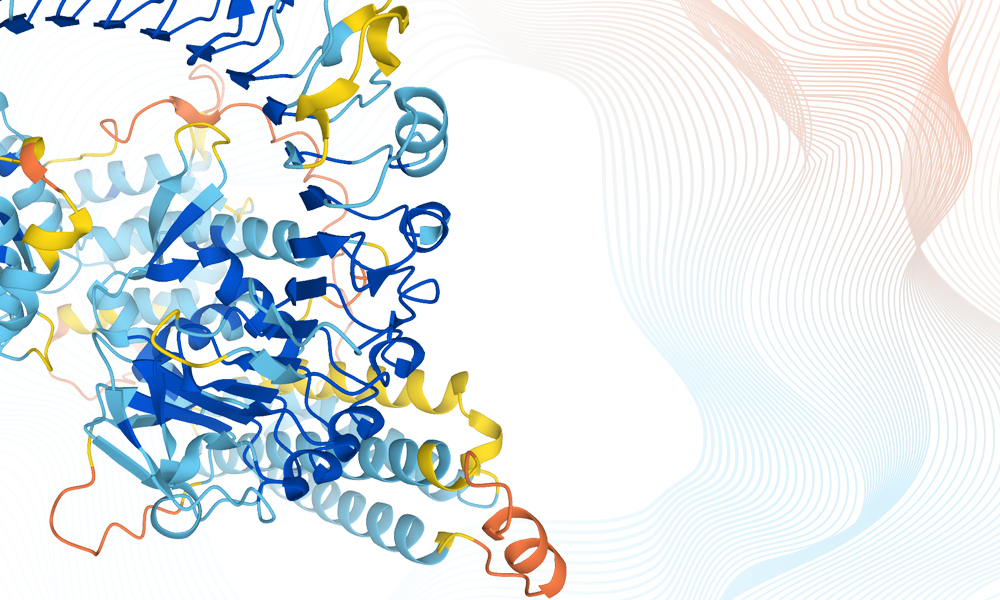
AlphaFold uses artificial intelligence to predict 3D protein structures. At the end of 2020, the CASP community recognised it as the first AI system to reach a level of accuracy similar to experimental models. In response, the scientific community called for DeepMind, whose scientists designed the AlphaFold system, to make the data and the computer code openly available.
DeepMind has now risen to the challenge. In collaboration with EMBL-EBI, it has made the AlphaFold protein predictions, source code and methodology freely and, crucially, openly available to the global scientific community through the AlphaFold database. The initial release contains more than 350,000 protein structures, from human and other species of biological interest, and this will expand to millions of proteins in the coming months.
Building on decades of expertise in making the world’s biological data available, EMBL’s European Bioinformatics Institute (EMBL-EBI) is working with DeepMind to ensure the predictions are Findable, Accessible, Interoperable and Reproducible (FAIR) so that researchers everywhere can make the most of them.
AlphaFold was trained using data from public resources – including UniProt, PDB and MGnify, which are co-hosted at EMBL-EBI – so it’s very fitting that its predictions are now openly available to all. This is a perfect example of the virtuous cycle of open data. By sharing data, the community can drive discovery faster than any one individual. Open data benefits all: public and private, experimental and computational, basic and applied research.
This ability to predict protein structure with unprecedented accuracy will underpin a revolution in biology as it allows us to understand better how all living things work. AlphaFold has many applications relevant to human health, agriculture and climate change.
By providing high-quality 3D structures for almost all human proteins, AlphaFold also frees structural biologists to focus their work on the more exciting questions of how proteins interact and function – something that AlphaFold doesn’t currently predict.
Enzymes, which are also proteins, are nature’s catalysts, but they are very difficult to design in a lab. Protein structure predictions can help scientists to design new enzymes, with new functions, such as processing waste or degrading plastics. Accurate protein structure predictions can also pave the way to improving crops so that they can handle climate change.
The possibilities for applications related to human health are endless, for example tackling some of the most serious diseases by predicting the structures of the proteins involved, characterising how they interact, and understanding how they cause disease. New proteins could be designed for novel vaccines or biological therapies to modulate diseases, and new candidate drugs can be identified more effectively.
Experimental researchers will be able to accelerate their structural studies to focus on complex biological systems, where experimental structural data at very high resolution are difficult to obtain.
While it’s true that AlphaFold is, so far, the gold standard for protein prediction, there are limitations to the method and the database, and these are important to note.
Almost all proteins function by interacting with other proteins, nucleic acids (DNA or RNA) or small molecules. AlphaFold doesn’t currently predict such complexes.
Proteins are also dynamic systems, with disordered regions that adapt their structure to their environment. Their dynamics and folding ‘from scratch’ have yet to be elucidated.
There are certain protein regions where AlphaFold produces only a low-confidence prediction (often for disordered regions). The AI system provides a confidence score as a helpful guide. Furthermore, AlphaFold has not been trained for predicting the effect of mutations, which can be critical in understanding why some individuals are susceptible to certain diseases. So like any method, AlphaFold will have its limitations that will inspire new and exciting avenues of research.
AlphaFold has illustrated the power of AI to improve 3D protein structure predictions. It complements existing methods and reveals new insights, but does not replace experimental methods to determine structures. This work serves as an exemplar of what is possible – and it is clear that AI will find many such applications in broader scientific research.
The power of AI underlies the AlphaFold predictions, based on data gathered by scientists all over the world during the last 50 years. Making these models available will undoubtedly galvanise both the experimental and theoretical protein structure researchers to apply this new knowledge to their own areas of research and to open up new areas of interest. This contributes to our knowledge and understanding of living systems, with all the opportunities for humanity this will unlock.
This post was originally published on EMBL-EBI News.
Nature 22 07 2021
10.1038/s41586-021-03819-2
Nature 15 07 2021
10.1038/s41586-021-03819-2