Read the latest Issue
MAPLE: a phylogenetic tool for pandemic-scale genome data
EMBL-EBI researchers have developed a new tool capable of performing state-of-the-art phylogenetic inference on larger datasets than previously thought possible
With the huge abundance of genomic data generated from life science experiments, processing large datasets remains a challenge in the field of bioinformatics. During the COVID-19 pandemic, the limited capabilities of existing bioinformatics tools meant that large amounts of data could not be analysed all at once, limiting the scope of evolutionary and epidemiological analysis.
To address this problem, a team led by researchers at EMBL’s European Bioinformatics Institute (EMBL-EBI) has developed a new bioinformatics tool that can handle large-scale genomic datasets, allowing scientists to analyse millions of viral genomes all at once.
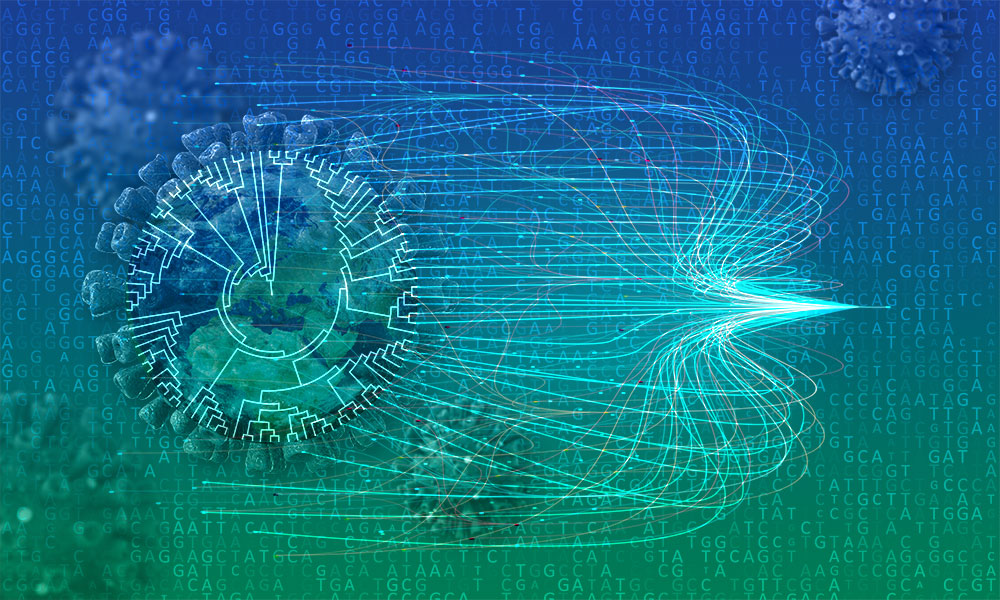
This research, published in the journal Nature Genetics, describes a new method – MAximum Parsimonious Likelihood Estimation (MAPLE) – that uses new mathematical approximations to develop an algorithm that works specifically on closely related genomes. This new approach enables rapid reconstruction of phylogenetic trees – a crucial step for understanding viral evolution and epidemiological spread.
Lessons learned from the pandemic
During the COVID-19 pandemic, researchers struggled to analyse the large number of genomic datasets generated. This made it challenging to study how the SARS-CoV-2 virus was evolving and spreading. Limitations of standard bioinformatic tools forced researchers to focus only on a small subset of samples at the time. Researchers everywhere soon realised that they needed faster and more efficient methods.
“We faced many challenges for analysing all the data that was coming in during the pandemic,” said Nicola De Maio, Research Staff Scientist at EMBL-EBI. “Traditional phylogenetic tools became inadequate as the data volume increased. We worked with others to try to ‘stretch’ these methods. We tried using supercomputers to solve the problem, but at some point, nothing seemed to work anymore. This prompted us to create MAPLE.”
The most significant advantage of MAPLE is its ability to process large-scale genomic data sets; millions of microbial genomes can be analysed at once.
Tools for epidemiological problems
Often, the tools used for studying evolution are the same whether you are looking at recent outbreaks of viruses and bacteria or at the evolution of distantly related species. To speed up phylogenetic inference within genomic epidemiology, the researchers developed a new algorithm that worked better for closely related samples – for example, viral genomes with only dozens of nucleotide differences, as is the case for SARS-CoV-2 genomes.
The researchers also realised that the lessons learned during this pandemic will be useful for bioinformatics tools moving forward. To be prepared for future pandemics, bioinformatic tools must cope with even larger scales of data.
“We as bioinformaticians learned a lot from the COVID-19 pandemic, but we also need to think about the future and how we can be better prepared,” said Nick Goldman, Group Leader at EMBL-EBI. “Bioinformatic tools need to be able to cope with more data, and we need tools for a range of specific tasks. New tools such as MAPLE can be a valuable addition to the bioinformatics community’s arsenal, helping researchers to process viral data faster and more efficiently for evolutionary analysis.”
Funding
This work was supported by EMBL core funding.
Source article(s)
Maximum likelihood pandemic-scale phylogenetics
Nature Genetics 10 April 2023
10.1038/s41588-023-01368-0
Want to track pandemic variants faster? Fix the bioinformatics bottleneck
Nature 1 March 2021
0.1038/d41586-021-00525-x