
Looping the loops: how chromosomes form
EMBL researchers and collaborators take important steps in unravelling chromosome formation
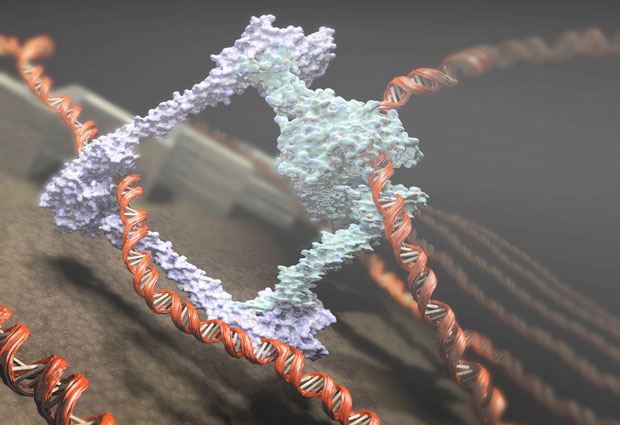
The nucleus of a human cell is less than one hundredth of a millimetre across but the DNA inside it, if stretched out, would be around two metres long. To fit into such a small space, the DNA has to be carefully coiled and packaged with various proteins. During cell division, the DNA molecules – in paired copies – are compacted still further, giving rise to the chunky X-shaped structures that are probably the most familiar manifestation of our chromosomes. How this compaction process happens is mysterious, but one important player is a ring-shaped protein complex called condensin. Two recent studies involving EMBL researchers have revealed important features of the condensin complex and its role in shaping chromosomes.
“Scientists realised about twenty years ago that condensin is a key organiser of chromosome structure,” says EMBL group leader Christian Haering, “but exactly how it works, nobody knows.” To play a role in organising DNA, condensin must have some way of grabbing hold of the DNA molecule. But, curiously, condensin doesn’t have any of the typical structures seen in other DNA-binding proteins. Haering’s group has now solved this puzzle by using X-ray crystallography to determine the structure of a central part of the condensin complex, both in the presence and in the absence of DNA.
Safety belt
The structure of DNA is like a spiral ladder. The rungs of the ladder are formed by the bases – the As, Cs, Gs, and Ts of the genetic sequence – and the uprights are formed by two chains of sugars and phosphates known as the sugar–phosphate backbone. The Haering group’s study, published in Cell, reveals a groove in the condensin complex that accommodates the DNA helix and makes contact exclusively with the sugar–phosphate backbone, not with the bases. This explains how condensin can bind to any part of the DNA, unlike other proteins that only bind to a specific sequence.
In spite of this, the complex still binds to DNA quite weakly. “Even with the DNA sitting inside this groove, it can easily escape again,” says Haering. A closer look showed that one of the proteins that makes up the condensin complex – called kleisin – encircles the DNA to keep it in place. “It creates a sort of safety belt, keeping the DNA buckled inside this groove,” Haering explains. “This is completely new – there’s no known protein that interacts with DNA in a similar manner.” When they modified the safety belt to prevent it from closing, they found that condensin does not associate with chromosomes any more.
Loop extrusion
One idea for how chromosomes could be compacted is known as the loop extrusion model. In this model, ring-shaped protein molecules interact with DNA, each one pulling a loop of DNA through its centre. It’s a little like taking a long thread and pulling loops of it through a backing material to make a carpet: very quickly the thread is packed into a much shorter space as it forms a series of loops rather than a long string. The problem is that no one has been able to show how any of the proteins that bind to DNA could form these loops. “We’ve seen how condensin holds DNA in this safety belt and how in principle the DNA could pass through the condensin ring to form a loop,” says Haering. “But that doesn’t explain how the loop could then be made larger and larger, as required by the loop extrusion model.”

Important evidence in support of this model has come from a second study, published in Science – a collaboration between Haering’s group and researchers at Columbia University in New York and the Kavli Insitute of Nanoscience Delft in the Netherlands. Together, they carried out a series of experiments in which they visualised the movements of condensin on long strands of stretched DNA. The condensin complexes travelled along the DNA strand over distances of several thousand DNA bases. “Similar protein complexes have been seen moving along DNA before,” says Haering, “but only passively sliding, not using energy to create this motor-like movement.” When the researchers added a second fluorescently labelled DNA molecule, they found that condensin was able to take this DNA molecule and move it in relation to the first. “We can imagine that condensin holds on to one side of a DNA loop and feeds the other side of the loop through the condensin ring, making the loop bigger,” Haering explains. “For the first time, we have experimental evidence showing how the loop extrusion idea could work in practice.”
Major steps
Previous experiments had suggested that condensin could not be the driving force behind loop extrusion, since its rate of energy consumption is too low. However, this conundrum can be solved if condensin is able to move in large steps along the DNA molecule. Figuring out exactly how condensin moves is therefore the next part of the puzzle. The condensin complex has two long protein units – called SMC2 and SMC4 – that might act like ‘legs,’ allowing condensin to walk along strands of DNA – each leg gripping the DNA strand in turn while the other leg moves forward. Another possibility is that the SMC units might stretch out to grab a section of DNA, then pull it in with a sort of scrunching motion, before reaching out for the next section. “These are plausible mechanisms to explain what we see in our experiments, but so far we’ve never seen any motor protein that can move with such large steps,” explains Haering. “It’s very unconventional – but that’s why trying to make sense of it is so fascinating.”


