
A physical revolution
EMBL physicist-turned-biologist alumni win 2017 Kendrew and Phillipson awards
By Margaux Phares and Sarah B. Puschmann
This year, two physicists-turned-biologists were given alumni awards for achievements in life sciences from EMBL: Matthias Mann, who was awarded the Lennart Phillipson Award, and Philipp Keller who won the 2017 John Kendrew Award. As their work demonstrates, physics informs biology – and across generations.
The path to identifying proteins
Mass spectrometry (MS) enables researchers to identify molecules such as proteins and nucleic acids quickly and accurately. Yet while the technique has been used by physicists for the past century, it was not until the 1990s that biologists began to make use of its potential. One of the people driving this biological revolution was EMBL alumnus Matthias Mann, a physicist who led the development of methods that helped bring MS from the margins to the mainstream.
“At the time, not many biologists considered using this technology in their research. And even if they did, they never thought it could actually work,” Mann recalls.
Mass spectrometry, a technique for sorting unknown molecules by their mass and electric charge in order to identify them, requires the molecules to be both electrically charged and in the form of a gas. In order to get them into this state, Mann’s former advisor John Fenn, developed a technology called electrospray ionisation, a method for producing charged particles, for which Fenn was awarded the Nobel Prize. Mann then took his advisor’s method one crucial step forward.
Electrospray ionisation requires a quantity of sample that, in the case of samples containing proteins and amino acids, often isn’t available. Along with his group, Mann found that by decreasing the size of the tip through which the solution is sprayed and lowering the rate of the liquid flow through the needle, this new technique, called nanoelectrospray, could be used even on biological samples in tiny quantities.
Still, nanoelectrospray had its limitations—it could only be used to identify peptides with different masses. (Peptides, like proteins, are compounds composed of amino acids arranged in a set order.) To distinguish peptides of the same mass, he and his team developed yet another technique. They found that peptides could be identified by determining only a tiny stretch—a few amino acids long—plus the masses of the surrounding amino acids. This characteristic chunk provided researchers with a unique fingerprint for the peptide, which they could then compare in a database to determine the identity of the peptide.
Mann’s postdoctoral student, Andrej Shevchenko, further improved sensitivity of identifying peptides by developing a technique to prepare samples containing proteins for nanoelectrospray. By staining the proteins using silver rather than the standard dye, the proteins of interest could be distinguished to a greater degree of sensitivity from the wash of background proteins in the sample.
These three achievements have made possible his and other teams’ identification of key proteins, which are key contributions to the field of proteomics, the mapping of all proteins in a particular organism. Recently, Mann, who currently works at the University of Copenhagen Denmark and at the Max Planck Institute (MPI) of Biochemistry, contributed mass spectrometry measurements to a study in which his team identified the almost 11,000 proteins contained in a healthy human heart.
We knew that this was a start for using mass spectrometry in biology.
As Mann looks ahead to identifying proteins in other human organs, his early work at EMBL continues to resonate with him.
“When we were able to find proteins even in minute samples, we knew that this was a start for using mass spectrometry in biology,” Mann recalls. “That was the best time.”
Fish development unmasked
Philipp Keller’s fascination with animal development dates back to his training as a physicist. “I enjoy understanding how complex systems work,” he says. “Some of the most complex things, you will find in biology – like a developing embryo or a higher functioning brain.” After studying physics and computer science at the University of Heidelberg, he went on to complete his PhD at EMBL. Not only did he have the opportunity to combine physics and biology studies during his PhD, but he also was able to work in three different labs and with three different advisors – an interdisciplinary experience that helped direct and shape his research.
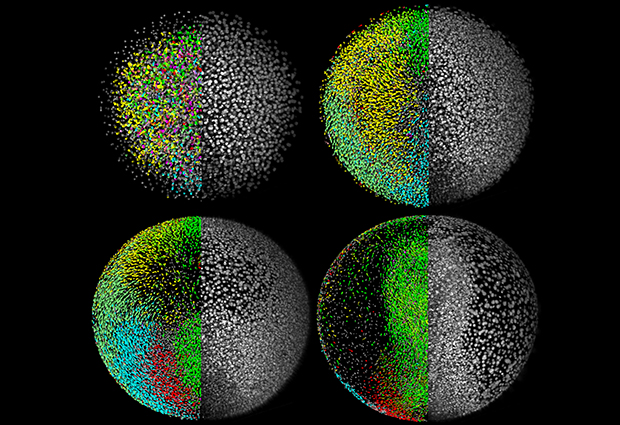
First Keller worked in Joachim Wittbrodt’s group on a project focused on visualising and reconstructing how a zebrafish forms from a single cell. They wanted to know how the single cell divides, migrates and develops into other tissues to form an embryo. It had never been seen before and it was technically difficult to study a vertebrate embryo at the cellular level. “It was a technology problem,” Keller explains, “so it was natural to form a collaboration with a group that was looking into new tools. I think EMBL is set up to encourage that.”
Keller then began work in Ernst Stelzer’s lab on a new kind of microscope, which he called the Digital Scanned Laser Light-Sheet Microscope (DSLM). While the group awaited some parts Keller needed in order to build the new microscope, Keller joined a third project headed by group leader Michael Knop: spore formation in yeast. “From a modelling perspective, it was interesting to me,” Keller says. “We wanted to understand this process through computer simulation in addition to experiments.”
I could see the developing fish right in front of my eyes!
Once the parts to create the DSLM in Stelzer’s lab arrived, Keller found that the skills he’d learned in computational modelling came full circle in the zebrafish project. The DSLM was faster and produced higher quality images than existing microscopes, which made it possible to image a developing zebrafish embryo for the first time.
“Rather than trying to work through a bunch of text and numbers, I could see the system – the developing fish – right in front of my eyes!”, Keller says. This gave him better insight into how biologists think about development at the cellular level.
After Keller completed his PhD at EMBL, he set off to map complex systems in the body as a group leader at the Janelia Research Campus of the Howard Hughes Medical Institute in Virginia, USA, where he is currently based. He continues to research developmental biology, in particular the nervous system and how it develops and first starts to function in the course of embryogenesis. “It’s not only important to know where the cells are located and what shape they have,” he says, “but it is also important to know what they can do – like how they become active, how they fire for the first time and how they interact with other cells to form complex circuits.” One example is looking at how activity of neurons in certain regions of the brain changes as we execute a certain behaviour, like walking or stopping.
But in order to see such minute and speedy activity, which can be measured by monitoring calcium concentrations in the neurons, his lab needs sensitive microscopes. Neurons are flooded with calcium when they fire, and they are emptied when they reset. It is possible to detect this activity with an indicator sensitive to calcium concentrations or even by measuring the electrical signals themselves, but neurons can spike one hundred or even one thousand times per second. “If you want to get the timing relationship right between cells, you need to match these timescales,” Keller explains. “This is why we want the microscopes to be as fast as possible, and this is why we are building a new generation of them.”




