ARISE2 has received funding from the European Union’s Horizon Europe’s research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 101178241.
ARISE2
Career Accelerator for Research Infrastructure Scientists
Overview
ARISE2 targets technology and method development experts with STEM backgrounds who are interested in using their expertise to advance research in the life sciences.
To address urgent challenges to human and planetary health new technologies and services are needed in the Life Sciences. ARISE2 is designed to meet this need by training fellows in the development of technologies and methods for improvement of scientific services, while also preparing them for careers in research infrastructures (RI) to make these advances available to the scientific community accelerating progress.
ARISE2 is an MSCA-funded postdoctoral fellowship programme offering talented STEM (science, technology, engineering and mathematics) fellows from around the world the unique opportunity to work on the development and/or improvement of technologies for Life Science Research while developing the expertise needed for a career in research infrastructures making them sought after experts both in academia and industry.

About EMBL
With 29 member states, laboratories at six sites across Europe and thousands of scientists and engineers working together, the European Molecular Biology Laboratory (EMBL) is a powerhouse of biological expertise. EMBL is an intergovernmental organisation with the mission of promoting molecular biology research in Europe, training young scientists, and developing new technologies.
Please watch the recording from the information webinar to learn more about the ARISE2 programme and how to apply:
Programme
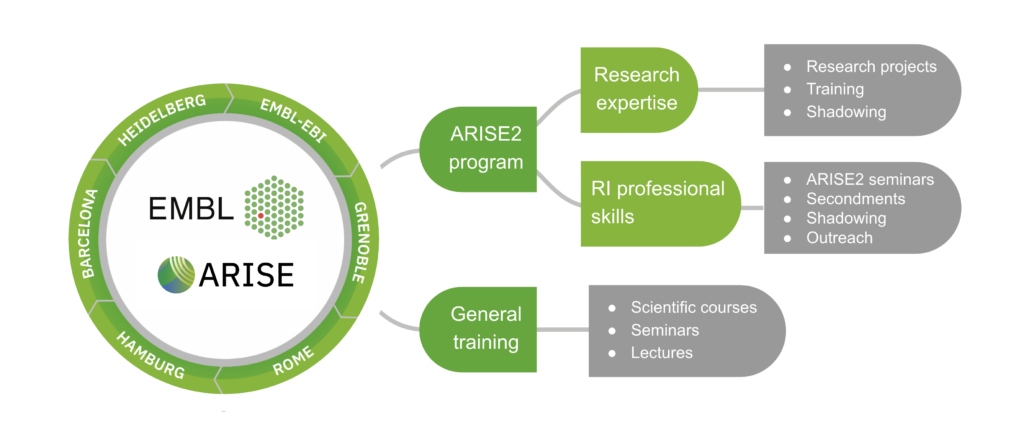
ARISE2 is a unique fellowship programme designed to train the next generation of Research Infrastructure (RI) experts through cutting-edge technology development, interdisciplinary collaboration, professional skills training and career support.
ARISE2 fellows receive 3-year contracts to work on the development of new technologies for the life sciences or the improvement of existing technologies. This is coupled to a comprehensive training programme that encompasses both programme-specific and individually tailored training opportunities and career development support. Training is divided into three core areas:
1. Research training including practical experience in service provision and job shadowing;
2. RI professional competencies;
3. Career development support and individualised training.
1. Research training including practical experience
Research expertise
Fellows work on self designed research projects to develop technologies for use in life science research. They optimise their technologies by applying it to the researcher projects of others gaining insight into different areas of life sciences, service provision and user needs. They prepare and maintain a data management plan for their projects according to EMBL’s Open Science Policy with the goal of managing their data according to FAIR and Open standards.
Supervision set up
Fellows have a main supervisor from EMBL, who is a group or team leader. Fellows also have an external co-supervisor for scientific consultations, either from the long secondment host organisation or a collaborating partner who participates in the 3 year research project. Finally, fellows have an academic mentor for career development consultations. It is also possible for fellows who are in research groups to have an advisor who has experience with service provision.
Practical Experience: Service Provision and Secondments
In line with ARISE2’s focus on hands-on training, fellows have the option to participate in the provision and maintenance of advanced services. This experience equips them with a deeper understanding of how RIs operate and the processes involved in service delivery. To allow fellows to focus on their own research project, this activity is not mandatory and limited to 10 days/year. Fellows hosted in groups that do not provide a service have a service-providing advisor to enable such an experience.
Fellows also complete secondments and short visits to enhance their interdisciplinary and intersectoral experience and support their scientific training and career development. These include two external secondments and two internal visits, enabling knowledge transfer across different sectors and expanding fellows’ professional networks.
| Research-supporting secondments |
|---|
| Long secondment (between 2-6 months) at a partner organisation. The partner collaborates on a part of the research project providing access to novel methods, instrumentation, and expertise. If the partner is collaborating on the full 3-year research project, fellows may spend up to 11 months on secondment. |
| Shadowing of a user at EMBL (5 days in the first 6 months of the fellowship): Fellows gain insight into how users prepare for service access, where and when they need support, their expectations, and how they handle data. |
| Career-supporting secondments |
|---|
| Short visit/job shadowing (1-2 weeks at any time in the fellowship) at a partner or non-partner organization: Fellows gain insight into the operational set ups at other organizations, develop professional skills, and build their networks. |
| Interdisciplinary visit to another EMBL facility (1 week at any time during the fellowship): Fellows learn about provision of service or technology development in complementary disciplines |
2. RI professional competencies and individualised training
RI professionals’ competencies are developed through a structured curriculum of mandatory ARISE2 seminars and a one-week school. This is reinforced during secondments, visits, pilot service provision of fellow’s technology and practical exercises in service provision (see 1) providing fellows with the skills needed to transition into senior roles within RIs.
The curriculum covers:
- Basic science policy relevant to RI
- Data Science including FAIR and Open research principals
- Service provision and user support
- Communication and outreach
- RI management (including budgeting, costing, impact assessment)
- Technology transfer and entrepreneurship
- Management of projects and people
The one-week ARISE2 school in the 2nd year of the fellowship focuses on strategic topics such as RI operations, team management, budgeting, defining and promoting services, collaborating with industry, and leveraging novel developments in RIs.
Outreach and Communication Training
ARISE2 encourages fellows to engage with the broader community through outreach and communication activities. Fellows participate in training provided by EMBL’s Science Education and Public Engagement office (SEPE) and Communications team, learning how to effectively promote the importance of life sciences and RI careers. They will take part in activities like public lectures, guided tours, and media interviews, helping them hone their communication skills.
3. Career development and individualised training
Each fellow creates a Career Development Plan (CDP) tailored to their long-term aspirations, supported by the ARISE Competency Framework for RI-specific skills and the ResearchComp from the European Commission for broader research competencies. Fellows meet with an EMBL’s Career Advisor early in the programme to assess their strengths and training needs and then periodically over the course of the 3-year fellowship. The CDP is reviewed annually to track progress and ensure ongoing alignment with career goals. The EMBL Fellows Career Service also provides guidance on training options, application materials, and workshops that facilitate the transition to their next professional role.
ARISE2 fellows are part of EMBL’s postdoctoral programme with access to additional training needed for their individual scientific and transferable skills development. EMBL provides a wide range of learning opportunities for its staff, including mandatory training in research integrity and data protection, a programme for complementary skills training for scientists and a Professional Development and Training programme.
ARISE2 Fellows
2024 Cohort

Disha Tandon
Postdoctoral Fellow (ARISE2)
Finn Group
EditJulian Gurgo
Postdoctoral Fellow (ARISE2)
Light Imaging Facility
EditProject: Microbial Variant Insights Knowledge Base (MVIKB): Mining, Collating, Mapping and Sharing Genetics and Epigenetics Variant Data in Microorganisms
Project: Combined genomics and transcriptomics imaging approach
“Imaging tools have revolutionised the field of spatial biology in recent years, particularly in spatial genomics and transcriptomics. Combined DNA and RNA labelling techniques are promising, as they allow exploring the role of 3D chromatin organisation in gene expression in both healthy and diseased organisms. My ARISE2 project focuses on developing a combined spatial genomics and transcriptomics imaging system, along with the automation of experiments, image processing, and analysis tools required to perform these experiments.”
Partner organisation: SciLifeLab
Maria Carla Carisì
Postdoctoral Fellow (ARISE2)
Hackett Group
EditNermin Akduman
Postdoctoral Fellow (ARISE2)
Microbial Automation and Culturomics Core Facility
EditProject: Developing Epigenome Editing Technologies for In-vivo Therapy
Project: Unlocking Microbial Diversity and Function through Automated Strain Isolation and Genome-Wide Profiling
“My ARISE2 project (Automating Microbial Isolation to Unlock Microbial Diversity and Function) is being developed at the Microbial Automation and Culturomics Core Facility. The project focuses on creating AI-guided, automated workflows for microbial isolation and characterization. By integrating artificial intelligence with robotic systems and high-throughput screening, it aims to accelerate the discovery of novel microbes and uncover their ecological and functional potential.“
Santiago Lago
Postdoctoral Fellow (ARISE2)
Stegle Group
EditSophia Bazzi
Postdoctoral Fellow (ARISE2)
Schneider Group
EditProject: Satellite-Powered Artificial Intelligence for eCosystEms biology – SPACE
Project: Development of an Algorithm for Automated Identification and Refinement of Hidden Biochemical Patterns in Protein Structures
Sotiris Samatas
Postdoctoral Fellow (ARISE2)
Sharpe Group
Edit
Yonathan Yitshak Goldtzvik
Postdoctoral Fellow (ARISE2)
Protein Data Bank in Europe
EditMicromechanics of Living Tissues across Scales
High throughput prediction of human protein complex structures
Eligibility
Applicants should be able to demonstrate prior experience in technology or method development, in the academia and/or non-academic sectors, relevant to the research fields of EMBL Research Infrastructures and services.
Academic requirements:
Applicants must hold a Ph.D. at the time of the call deadline (September 30, 2025 for the 2025 call). Researchers who have successfully defended their doctoral thesis but who have not yet been formally awarded the degree are eligible to apply. Successful candidates have 4 months to take up their fellowship.
Mobility requirements:
The programme is open to experienced researchers from around the world who are interested in the development or improvement of technologies to support life science research. Prior association (including visitor contracts) of an applicant with EMBL is compatible with application to the programme but cannot exceed 12 months within the last 3 years prior to the application deadline. Prior association relates to having worked with a person/group. It can include having done a Ph.D. or Postdoc with the supervisor or having been a visitor in their lab/group. Any previous association must be indicated on the application form.
Programme requirements:
Applicants must use the application form and project template, including ethics check form, available in the “How to apply” section for their applicant to be eligible. At least one reference (up to 3 possible) is due by the call deadline.
If you have questions related to your personal situation please get in touch with us at arise2@embl.org.
Apply
Application
How to apply
1) Read the guide for applicants and fill in the application form:
2) An information session about the programme and how to apply recently took place. Please watch the recording to learn more about the ARISE2 fellowship and application procedure.
3) Projects: You are invited to propose a project of your own design which relates to the development of new or improvement of existing methods or technologies and which can be applied to the scientific questions of other researchers as a service and integrated into Research Infrastructures. The proposal should, whenever possible, foresee how the developed technology will simplify or even automate FAIR data management for users, throughout the data life cycle. The project should extend beyond local interest, having potential for international transfer or user base.
Please note — Projects should use the application template provided below, which includes the Horizon Europe ethics self-assessment on the project:
4) Projects require an EMBL supervisor and can include external collaborators. Please get in touch with the EMBL group leaders participating in the call who you are interested in to discuss your project idea.
5) Secondments: Part of your training involves a secondment with an ARISE2 partner organization or other external partner of between 2-6 months (or up to 11 months if the second host is participating in your project). The purpose of the secondment is to provide access to novel methods, instrumentation and skills. As part of your application you may propose a secondment host (see list available in the annex of the Guide for Applicants) but this is not mandatory at this stage. Please note that you can change your choice of secondment host up until the 6th month of your fellowship if your needs and that of your project change.
6) Open an online application:
Your completed ARISE2 application form (see 1 above) and project with ethics prescreen (see 3 above) should be converted to a single pdf and uploaded in the CV and coverletter section of the online application.
For your application to be eligible the following is required:
- You must meet the programme’s eligibility requirements.
- Your application must be submitted via the application portal (available here).
- We must have at least one (up to 3) reference by the call deadline (September 30, 2025). Please ask your referees to send their reference to ARISE2-references@embl.de
- Your proposal must use the provided template and you must also complete the Horizon Europe ethics self evaluation on your project (see 3).
Evaluation of submitted applications
ARISE2 is a competitive merit-based fellowship programme. It follows a well-defined weighted scoring system (see the Guide to Applicants). Eligible applications are independently reviewed by 3 external experts. Evaluators are asked to provide an overall impression of a candidate’s application in terms of excellence, impact and implementation as described in the Guide to Applicants.
Interviews
Lab visits: Interviewing candidates interact with the lab(s) involved in their proposed projects prior to the interviews.
Interviews: The interviews for the 2025 ARISE2 call will take place during the week of December 1st, 2025 by Zoom. Candidates are asked to give a presentation on their research proposal and career achievements (20 minutes). This will be followed by a a panel interview (25 minutes).
Appeal
Applicants who are ineligible, not short-listed or who don’t receive an offer after interviews can appeal the decision if they feel a procedural error limited the success of their application. The scientific decisions of the Evaluators and the Interview Panels are not open to appeal. The process for appealing is included in the outcome email to applicants.
If you have question regarding the Programme and application procedure, please contact us at arise2@embl.org.
Participating hosts
The group leaders participating in the ARISE2 programme, their areas of interest and current and future work plans will be available below, and in the guide to applicants from July 1st, 2025. Please read this material before contacting the group(s) you are interested in to discuss your project ideas. For contact details click on the names of the group leaders.
Data and computational sciences
The groups listed here are working on the development of technologies in AI and machine learning, software development, bioinformatics and computational modelling, data sciences, including data management, data science, big data, data standards, information retrieval and relevance ranking.

Alex Bateman
Senior Team Leader – Protein Sequence Resources
EditResearch interests
The sequence families team develops the InterPro, Pfam, Rfam and RNAcentral databases. This provides and excellent environment to learn and help develop cutting edge information resources. At present we are rapidly adopting AI technologies to assist in coding and curation. There are many opportunities and we are increasing our ambitions to provide detailed and accurate biological information more rapidly to the community. Projects could focus on using AI to improve data, or the software infrastructure for any of the data resources we provide.

Ewan Birney
Interim Executive Director, EMBL
EMBL-EBI Directors Office
EditResearch interests
We have developed a framework to allow genome wide association testing for CNVs to be performed at scale in large human cohorts and have developed a set of guidelines for standardisation of CNV GWAS models. We will work closely with the GWAS catalog to implement infrastructure that is acutely needed to allow the long term capture of CNV GWAS results under FAIR guidelines and the linkage of information across variant types (i.e. SNPs and CNVS). We will continue development of CNV GWAS methods and also apply the existing standard CNV GWAS models to large human cohort data. This will generate larger volumes of CNV GWAS results that can be irectly deposited into the GWAS catalog and will drive the development of infrastructure for data linkage and additional CNV specific tooling and functionality within the GWAS catalog.

Clement Blanchet
Team Leader
EditResearch interests
Our team provides comprehensive support for small-angle X-ray scattering (SAXS) experiments, covering experimental design, setup, data collection, analysis, and interpretation. We focus on developing advanced SAXS methods, including time-resolved and high-throughput approaches, while continually improving the beamline infrastructure and sample environments. We also work on enhancing data reduction and analysis tools, with an increasing emphasis on simplifying and automating FAIR data management for users. Fellows will have the opportunity to engage in experimental innovation, computational development, and service provision within a collaborative and multidisciplinary environment.

Emily Clark
Genome Analysis Team Leader
EditResearch interests
The genome analysis team develops state-of-the-art cloud-based data analysis and portal infrastructure to coordinate, analyse, enrich, and present the wealth of genomic and phenotype data arising from global agricultural and biodiversity projects. This includes projects such as the EuroFAANG Research Infrastructure Project that aims to create a sustainable pan-European infrastructure for genotype-to-phenotype research across farmed animal species. We are continually seeking projects that utilise cutting edge cloud data engineering to design and develop analysis, visualisation and data management infrastructure at significant data scale. This could include development in the areas of pangenomics, single cell atlases, cloud-based interactive analysis platforms, generating new bioinformatic analysis workflows, integration of genomic and phenotype data and ‘omic data visualisation. Data analysis platforms and portals are crucial services to enable and accelerate global agricultural and biodiversity research, tackling key societal issues of food security, climate change and biodiversity loss.

Gautam Dey
Group Leader
EditResearch interests
Research in my group is focussed on the evolution and diversity of mitotic mechanisms across the eukaryotic tree of life. Projects incorporate an interdisciplinary wet- and dry-lab toolbox that includes comparative genomics, quantitative cell biology, imaging, and experimental evolution in multiple microbial model species. In order to answer our long-term research questions, we often find ourselves in the position of having to improve existing tools (for example, our recent work on adapting ultrastructure expansion microscopy for fungi: see Hinterndorfer, Mikus, Laporte et. al. 2022, Reza et al. 2024, Mikus, Shah, Rubio 2024) or develop entirely new technologies, with a commitment to disseminate and teach as broadly as possible. Future technology development projects: 1. Scalable U-ExM experimental and data management pipeline for ultrastructural atlases (of marine plankton, related to TREC, Moore Foundation and EMBL Planetary Biology projects); cross-modality integration with volume EM techniques (experimental and computational – AI and machine learning, bioinformatics, software) 2. Structure-aware comparative genomics and proteomics methods for investigating deep evolutionary homology (computational – bioinformatics, software) 3. Developing an image-enabled phenotypic selection platform for microbial experimental evolution (building on the work exemplified by Helsen et al. Nature Cell Biology 2024) (experimental and computational – AI and machine learning, engineering)

Robert Finn
Section Head, Team Leader and Senior Scientist
Microbiome Informatics
EditResearch interests
Our team specialises in the assembly and annotation of microbial genomes, whether derived from isolates or complex communities. As we broaden the taxonomic diversity of studied microbes—including viruses, prokaryotes, and eukaryotes—functional annotation becomes increasingly challenging. While most current approaches focus on annotating individual genes, our research aims to develop new methods that integrate large-scale and multi-omic datasets, as well as structural models, to predict higher-order biological features such as operons, metabolic pathways, protein complexes, and inter-species interactions.

Michael Dorrity
Group Leader
EditResearch interests
The Dorrity lab develops new technologies to for statistical developmental genetics, focusing on (1) new molecular approaches for ultra-scalable multi-omic single cell profiling and (2) new computational methods and software to manage and analyze the multi-scale data resulting from these experiments. We are also collaborating with the “Frontier” Organism Center to generate systematic spatiotemporal perturbation datasets to learn how the environment affects cellular and organismal function. We have opportunities for potential ARISE fellows at each level.

Christina Ernst
Functional Genomics Team Leader
EditResearch interests
The Functional Genomics Team at EMBL-EBI develops core open-access resources for transcriptomics data reuse, including Expression Atlas and Single Cell Expression Atlas. We build the pipelines, metadata standards, and user interfaces that enable researchers worldwide to submit, access, and explore transcriptomic data from a wide range of organisms in line with FAIR principles We welcome project proposals that broadly align with our efforts to improve the accessibility, reusability, and scalability of transcriptomic data. Areas of interest include:
Extending infrastructure to support single-cell RNA-seq data from diverse and non-model species, particularly in collaboration with the Biodiversity Cell Atlas community;
Adapting to emerging technologies such as spatial transcriptomics;
Developing approaches that enhance automation in data processing and improve the AI-readiness of our resources.

Maria Marta Garcia Alai
Head of EMBL SPC Facility
EditResearch interests
Development of Software for X-ray Tomography of Sediment Samples We use X-ray tomography to explore and identify microbial ecosystems within sediment samples. The ARISE Fellow will contribute to data analysis, focusing on the visual identification and segmentation of various organisms such as diatoms and cyanobacteria. Our goal is to integrate machine learning models for automated extraction and classification, streamlining the analysis process and improving accuracy. Development of Software for Molecular Biophysics Our team is developing eSPC, an online data analysis platform for molecular biophysics (https://spc.embl-hamburg.de/). Each module follows a standardized workflow: data loading, parameter input, model selection and fitting, followed by graph production and data export. We aim to integrate AI-driven decision-making to enhance quality control, optimize model selection, and suggest follow-up experiments—streamlining analysis and reducing reliance on human expertise. This would not only improve data reliability but also broaden accessibility to non-specialists.

Matthew Hartley
BioImage Archive Team Leader
EditResearch interests
EMBL-EBI’s imaging databases, the BioImage Archive and EMPIAR (Electron Microscopy Public Image Archive) are used by tens of thousands of scientists, who provide and access life sciences images. They have strong interactions with both EMBL’s world leading imaging facilities, as well as the worldwide BioImaging community. This gives us the ideal position to develop image data conversion, storage, compression, visualisation and analysis technologies. All our technology development is primarily applied to improve our service offering to imaging scientists, image analysts and researchers in the biological sciences. Automation of FAIR data management, particularly through AI processes (e.g. LLM agents, MCP servers), and application to multimodal data (e.g. spatial transcriptomic) is a key strategic goal for our service development.

Henning Hermjakob
Head of Molecular Systems
EBI Molecular Networks
EditResearch interests
The Molecular Systems Team provides a broad portfolio of resources for systems biology, ranging from molecular interactions (IntAct) and curated pathways (Reactome) to systems biology models (BioModels) at the highest level of abstraction. Supporting the domain-specific resources, our team develops broad-scope data infrastructure resources identifiers.org, Omics Discovery Index, and EBI Search. Our services range from medium to heavy use (>100,000 users/month), and from small (a few GB installed size) to very large (indexing > 5 billion data objects across 40 major resources).
Our resources are recognised by their respective communities (5 x Elixir Core/Data Deposition/Interoperability and 2 x Global Core Biodata Resource recognition) and embedded into a framework of European and global collaborations.
All services are open source, open data, and emphasize content quality, stability, high availability, and user-friendly visualisation. We constantly evolve our services to provide more efficient data access and analysis in a context of rapidly growing data volumes. ML techniques are increasingly used for user guidance, data discovery, and content curation support, and we strive to evolve our datasets to be AI-ready in a rapidly changing scientific environment.

Alexandra Koumoutsi
Head of Microbial Automation and Culturomics Core Facility
EditResearch interests
The Microbial Automation and Culturomics Core Facility will provide access and training to cutting-edge equipment and knowhow related to automation for microbial work, with an initial focus on the human gut microbiome. Moreover, the facility will enable the deconvolution of microbial complex communities (isolation of most/ all species within a community). Using robotic systems inside/ outside of controlled environmental chambers, researchers will be able to construct and characterize genome-wide mutant libraries, assemble strain collections or communities and perform large-scale screens (from phenotyping to compound screens). The facility aims to create and provide genetic tools and resources for non-model species and microbial communities and establish analysis pipelines for genomic to high-throughput phenotypic data.
Research interests
We develop innovative, ethical and sharable approaches which help realise the power of sequencing to control pathogen threats. We are interested in both producing methods, and generating biological insights by applying these methods to data from a range of sources. We currently have two main lines of research: understanding the mechanisms of pathogen evolution, and developing methods and resources that lower the barrier to entry for infectious disease genomics and modelling. We have particular interests in fast k-mer search, improving genome annotation, and integrating different data sources to enrich microbial sequence data.

Julia Mahamid
Head of Molecular Systems Biology
Molecular Systems Biology Unit
EditResearch interests
In-cell structural biology attained via cryo-electron tomography (cryo-ET) of intact cells is currently at very exciting stage; as the highest resolving technique to image the unperturbed cellular machinery in three dimensions (3D), cryo-ET produces data that can be quantitatively analyzed to derive mechanistic models of heterogeneous macromolecular assemblies within their functional context. With exceptional access to state-of-the-art infrastructure at EMBL, scientific and industry collaborations, we address experimental and computational bottlenecks in our workflows, pushing the boundaries of possible applications and increasing their robustness to make our methods more widely available together with the service teams at the EMBL Imaging Centre. The increasing users demand for these technologies necessitates developments and automation, in particular in advanced cryo-plasmaFIB volume imaging and lift-out lamella preparations for vitrified multicellular specimens, assisted by 3D correlative image registration. The increasing data volumes that are already being generated demand robust solutions for data management and flow to the public data repositories at EMBL-EBI (EMDB, EMPIAR and BioImage Archive).

José Márquez
Head of Crystallisation Facility
EditResearch interests
Our Team has pioneered the development of Online Crystallography; fully automated protein-to structure pipelines integrating crystallization, synchrotron data collection and crystallographic data analysis into continuous workflows operated via the web. These pipelines are currently used by hundreds of scientists worldwide and are based on the CrystalDirect technologies and CRIMS software, which we have contributed to develop. Recently, we have implemented a fully automated pipeline for ligand and fragment screening to support structure guided drug design. EMBL Grenoble is co-located with the European Synchrotron Radiation Facility (ESRF) in Grenoble, which produces some of the world’s most brilliant X-ray beams worldwide. EMBL and ESRF jointly operate six crystallography beamlines one of which is the fully automated MASSIF-1 whose operation is highly integrated with the EMBL’s HTX Lab. Our interdisciplinary team offers opportunities for scientists, engineers and software developers to work in one of the leading infrastructures for structural biology within the areas of protein crystallography, structure-based drug design, automation, and large-scale scientific data management and analysis. Currently, we are particularly interested in profiles either in structural biology or computer science orientated towards one or several of the following areas: X-ray-based fragment screening, structure guided drug design, AI-based and advanced computational approaches for hit-to-lead optimisation, medicinal chemistry.

Fergal Martin
Eukaryotic Annotation Team Leader
EBI Eukaryotic Annotation
EditResearch interests
The team builds tools, pipelines and workflows to annotate genomes and subsequently release those annotations through the Ensembl genome browser. Specific area where development is occurring are: 1) Applying deep learning to gene prediction (both methods development and testing of emerging tools), 2) Pangenome annotation – how to accurately and quickly remap annotation from a reference species to a pangenome, how to integrate haplotype/breed/strain specific annotations to capture novelty, how to deal with CNVs and mismapping issues, 3) Better integration of multiple data streams to build a high-quality final gene set (maximising the available evidence to build a high-confidence set of transcripts, 4) The creation of training sets and standardisation of metadata and analyses across genomes (standardising things like short/long read mappings, protein-to-genome alignments and repeat analysis for use in downstream training), 5) Refactoring and optimisation of legacy annotation pipelines into modern languages and workflows, 6) Cost-benefit analyses of annotation processes (understanding quality versus compute cost and operational complexity), 7) Dashboards and metrics for monitoring annotation pipelines and metrics on their output, 8) Integration of Ribo-seq data and understanding the dark proteome (with a view to integration in Ensembl).

Noel O'Boyle
Chemical Biology Resources Team Leader
ORCID: https://orcid.org/0000-0003-4879-2003
EditResearch interests
Our team develops tools and resources that help record, analyse and interpret the biological effect of compounds, typically small molecules. The ChEMBL database of bioactivities extracted from the literature is widely used to support drug discovery in academia as well as throughout the pharmaceutical industry; ChEBI is an ontology and database of chemicals of biological interest whose identifiers are widely used by other biological databases; SureChEMBL is a database of molecules and biological targets extracted from the patent literature via a fully automated process. Our team is a mixture of researchers, data scientists, software engineers and data curators. The work within the group ranges from curation of biological assay descriptions and chemical structures, applying NLP and AI approaches to extract relevant information from unstructured text, through to web development with modern technologies. Team members benefit from working in an open and collaborative environment which in particular for an ARISE fellow provides the opportunity to grow both in knowledge of service provision and technology development. The close alignment of our services with the pharmaceutical industry will also be of benefit.

Gergely Papp
Team Leader
EditResearch interests
The Papp Team at EMBL Grenoble develops scientific instrumentation for macromolecular X-ray Crystallography and Cryo-Electron Microscopy applications. One of our most recent and significant development projects is the EasyGrid technology, a system designed to fully automate sample preparation and quality control for cryo-imaging experiments. Pilot measurements demonstrated that the EasyGrid system is suitable for preparing samples for Single Particle Analysis experiments and showed the superiority of EasyGrid-prepared samples in terms of ice quality for Cryo-Electron Tomography and X-ray Nano Imaging techniques, compared to traditional plunge freezing equipment. Based on these solid technical developments, the team is now working on creating software tools to fully automate the Cryo-EM/ET sample preparation workflow, including an AI-based system for preparation parameter optimization using an on-the-fly sample quality control method.

Fabio Petroni
Group Leader
ORCID: 0009-0005-5996-9830
EditResearch interests
I am a Computer Science researcher focused on Artificial Intelligence, particularly natural language processing, information retrieval, and machine reasoning. My work explores how machines can access, interpret, and synthesize knowledge at scale. I co-developed the Retrieval-Augmented Generation (RAG) framework and have contributed to foundational work on treating large language models as knowledge stores and reasoning engines. At EMBL, I aim to establish a novel AI-driven agentic system where humans and machines collaborate to accelerate scientific discovery by surfacing insights, identifying knowledge gaps, and formulating new hypotheses from large, multimodal corpora.
Fabio is collaborating with Ola Spjuth from SciLifeLab. They welcome projects that explore building Al agents that actively
support scientific discovery by navigating, synthesizing, and reasoning
over complex biological data. The goal is to create collaborative systems
that help researchers uncover insights and formulate new hypotheses
across disciplines.

Gerald Pfister
Head of Flow Cytometry Facility
EditResearch interests
A primary technology development focus at the EMBL Rome FCF is advanced flow cytometry and cell sorting. These tools are crucial for accurate single-cell data acquisition and for purifying cells for diverse downstream applications like RNA/DNA analysis and single-cell sequencing. Current projects include developing & optimizing flow cytometry protocols for a number of different samples & cell types. Bulk- and single-cell sorting assays of various different sample- and cell types are routinely designed and performed. Recently, we started to set up assays for soluble protein analysis by flow cytometry, and are exploring novel imaging-enabled flow cytometry assays. Currently, two high-end cell sorters (BD FACSAria Fusion with BSL-2 cabinet & BD FACSMelody), an Attune NxT flow cytometer with autosampler, and a 10X Genomics Chromium controller are available in the facility For ARISE2, the FCF, as the host lab and main technology development partner in advanced flow cytometry and cell sorting techniques, is seeking collaborative project proposals. These projects will leverage our cutting-edge capabilities with robust bioinformatics approaches. We are particularly interested in proposals that align with the biological research focus of our collaborator, the Rompani lab. This includes generating and analyzing complex datasets to explore molecular underpinnings of biological phenomena, such as those related to age-related changes or cellular profiling linked to higher-level phenotypic data. A strong bioinformatics component, centered on developing novel analytical tools and integrated interpretation pipelines, is essential for these projects. This integrated approach, emphasizing the creation of new analytical methods rather than mere application, will offer comprehensive data lifecycle experience, fostering technology creation.

Robert Prevedel
Group Leader and Senior Scientist
EditResearch interests
We are developing advanced optical imaging methods that are based on multi-photon microscopy, active wave-front shaping, photo-acoustics as well as high-resolution spectroscopy. Our aim is to establish our new approaches as disruptive technologies in the life sciences and to further engineer and automate our prototypes for routine service provision.

Julio Saez Rodriguez
Head of Research
Saez-Rodriguez Group
EditResearch interests
Our group has been developing methods for spatial omic data analysis, integration with prior knowledge and AI. We are interested in project proposals related to developing a platform for data sharing and analysis of raw spatial multi-omic data sets at EBI, incorporating core methodologies of our lab to analysis spatial multi-omic data, relying on the integration of curated prior knowledge to decipher underlying molecular mechanisms as well as developing dynamic models of cell-cell interactions and tissue reprogramming. SciLifeLab would also provide support to applicants and their projects.

Sinem K. Saka
Group Leader
EditResearch interests
We are at the forefront of developing cutting-edge technologies that extend the boundaries of spatial biology research. Our focus is on understanding how the molecular and spatial compositions of cells are linked to their phenotypes. To achieve this, we have established state-of-the-art methods for imaging-driven spatial omics (multiplexed protein / RNA detection, imaging-base ex situ omics analyses) and have automated workflows that seamlessly integrate hardware and software for adaptive feedback microscopy, in collaboration with ALMF. Additionally, we develop and disseminate detection assays (and the associated reagent chemistries) that enable multiplexed imaging, spatial barcoding and spatial omics methods. Experimental: We are interested in spatial biology and address it with super-resolution, multiplexed imaging and spatial omics technologies that we have developed using DNA nanotechnology. We aim to push custom spatial omics approaches with 3D implementation or non-model organisms, which will be unique and very useful to many groups at EMBL and beyond to investigate the heterogeneous ecosystem of different tissues, including reproductive/developmental biology, planetary biology, microbiome and exposome studies in human tissues and disease models with a wide range of sample types (organs-on-a-chip, thick tissue slices, whole embryos or organisms, organoids, organs, xenografts). We have multiple collaborations with academic groups and industry where we perform generalized application or custom adaptation of our methods, and hence have many use cases for the service provision. Computational / data-oriented: Additionally, we are building an expansive collection of multidimensional spatial data, which is particularly well-suited for novel AI and machine learning-based analyses. The analyses results can similarly drive generation of new targets sets and development of new assays, and we would have the capacity to embed method development in this feedback system between analyses, experimental design and data generation. The datasets we generate and work with represent next generation data types and modalities that are increasingly being generated and are anticipated to be represent popular future use cases for a wide range of data-related services. Our group is dedicated to exploring the potential of advanced data analysis methods on next-generation spatial data, characterized by its rich and complex feature space. This involves both the technology development aspect, where we innovate new analytical approaches, and the support of essential infrastructure, including data formats, metadata schemas, and FAIR data management practices. These elements are vital for both research and service provision.

Ugis Sarkans
Technical Team Leader
BioStudies Team
EditResearch interests
Our team builds and maintains the BioStudies database – a resource that facilitates transparent, reproducible science by aggregating and publishing all outputs of a scientific study. BioStudies acquires data via a variety of routes, both pre- and post-publication. We are looking to extend our infrastructure and enable application of data harmonisation methods to support new, emerging fields, while maintaining the generic nature of our services.

Christian Tischer
Team Leader
Data Science Centre
EditResearch interests
Volume electron microscopy (vEM) is a fast-evolving imaging field that aims at acquiring ultrastructural details of cells, small model organisms, and tissues in three dimensions. vEM generally produces large datasets that can reach multiple TB of data, for which extracting valuable morphometric information by image segmentation is still a challenge. For targeted studies, which restrict measurement to selected regions of interest, other imaging modalities (light microscopy (LM) or X-ray microscopy (XRM)) are used to build volumetric maps of the samples in which sub-regions of interest are defined. Due to the size and complexity of such multi-modal image datasets it is challenging to efficiently (ideally automatically) extract biophysical information from these information-rich datasets. To tackle this challenge, the Bioimage Analysis Support Team and Electron Microscopy Core Facility from EMBL Heidelberg as well as the VIB Bio Imaging Core from KU Leuven are teaming up to develop and deploy re-usable computational workflows for the automated management and (AI-based) analysis of such correlative volume EM datasets.

Juan Antonio Vizcaino
Team Leader, Proteomics & Metabolomics
EditResearch interests
Improving the functionality of the EMBL-EBI resources PRIDE and MetaboLights, and the integration of proteomics/metabolomics data with other omics data types (implementing the FAIR data principles) are two key aspects for the team in the near future. This offers the possibility for the fellow to work in different topics depending on their background (e.g. data analysis, data visualisation, software development and infrastructure, data management practises, Artificial Intelligence, etc). In the context of data integration for proteomics data, as mentioned above, this involves different data types such as gene and protein expression information (together with Expression Atlas and Open Targets), post-translational modifications (UniProt), (meta)proteomics data and (meta)genomics sequences (Ensembl, MGnify) and also structural proteomics data such as crosslinking (PDBe). In the context of metabolomics, integration of metabolomics with other data types (as mentioned above) and providing improved support for lipidomics data, the generation of public spectral libraries and interaction with ChEBI and other relevant EMBL-EBI resources are topics of high-interest.

Kyle Morris
EMDB Team Leader
Electron Microscopy Data Bank
EditResearch interests
The Electron Microscopy Data Bank manages the public repository for electron microscopy maps and representative tomograms of macromolecular complexes and subcellular structures. We develop technologies and services that facilitate open-access to published cryoEM data, add value by revealing facets of the data we hold, connections to complementary databases and we use our work to prepare the archive for future experimental technologies.

Andrew McCarthy
Team Leader
EditResearch interests
The McCarthy Synchrotron technology team at EMBL Grenoble operates a suite of modern structural biology beamlines with the ESRF. We continually develop new methods and automation routines for macromolecular crystallography and small angle X-ray scattering experiments. We have also started to develop biological X-ray imaging techniques in collaboration with the Papp team and our ESRF colleagues. In the context of this particular ARISE2 call we interested in proposals related to either: 1) the further development of advanced in situ serial room temperature crystallography techniques and 2) the development of biological X-ray imaging applications at synchrotrons.

Michael Zimmermann
Group Leader
Zimmermann Group
EditResearch interests
Our workflows are tailored to the diverse needs of metabolomics research, spanning targeted and untargeted metabolomics, lipidomics, and fluxomics. By automating key steps in data handling, we provide researchers with efficient, reliable tools to uncover insights into metabolic processes.
In line with EMBL’s commitment to Open Data, the MCF also contributes to global data sharing efforts by depositing processed data and results into MetaboLights at EMBL-EBI, one of the leading repositories for metabolomics research. This approach promotes transparency, reproducibility, and accessibility, enabling the wider scientific community to build on our work. Through these activities, the facility supports progress in fields such as systems biology, personalized medicine, and biotechnology by providing standardized data and state-of-the-art analysis methods.
Imaging and optical engineering
The groups listed here are working on development of technologies in imaging, microscopy, image analysis, X-ray optics, optical instrumentation developments.

Alvaro Hernan Crevenna Escobar
Head of Light Imaging Facility
EditResearch interests
A biology/physics group with expertise in microscopy, histology and image analysis for two purposes: a, to further develop imaging technology; and b, to establish complex platforms such as spatial transcriptomics, tissue profiling and correlative X-ray imaging/super-resolution microscopy. We aim at bringing these services for the wider European research community through academic or industry collaborations.

Gautam Dey
Group Leader
EditResearch interests
Research in my group is focussed on the evolution and diversity of mitotic mechanisms across the eukaryotic tree of life. Projects incorporate an interdisciplinary wet- and dry-lab toolbox that includes comparative genomics, quantitative cell biology, imaging, and experimental evolution in multiple microbial model species. In order to answer our long-term research questions, we often find ourselves in the position of having to improve existing tools (for example, our recent work on adapting ultrastructure expansion microscopy for fungi: see Hinterndorfer, Mikus, Laporte et. al. 2022, Reza et al. 2024, Mikus, Shah, Rubio 2024) or develop entirely new technologies, with a commitment to disseminate and teach as broadly as possible. Future technology development projects: 1. Scalable U-ExM experimental and data management pipeline for ultrastructural atlases (of marine plankton, related to TREC, Moore Foundation and EMBL Planetary Biology projects); cross-modality integration with volume EM techniques (experimental and computational – AI and machine learning, bioinformatics, software) 2. Structure-aware comparative genomics and proteomics methods for investigating deep evolutionary homology (computational – bioinformatics, software) 3. Developing an image-enabled phenotypic selection platform for microbial experimental evolution (building on the work exemplified by Helsen et al. Nature Cell Biology 2024) (experimental and computational – AI and machine learning, engineering)

Maria Marta Garcia Alai
Head of EMBL SPC Facility
EditResearch interests
Development of Software for X-ray Tomography of Sediment Samples We use X-ray tomography to explore and identify microbial ecosystems within sediment samples. The ARISE Fellow will contribute to data analysis, focusing on the visual identification and segmentation of various organisms such as diatoms and cyanobacteria. Our goal is to integrate machine learning models for automated extraction and classification, streamlining the analysis process and improving accuracy. Development of Software for Molecular Biophysics Our team is developing eSPC, an online data analysis platform for molecular biophysics (https://spc.embl-hamburg.de/). Each module follows a standardized workflow: data loading, parameter input, model selection and fitting, followed by graph production and data export. We aim to integrate AI-driven decision-making to enhance quality control, optimize model selection, and suggest follow-up experiments—streamlining analysis and reducing reliance on human expertise. This would not only improve data reliability but also broaden accessibility to non-specialists.

Matthew Hartley
BioImage Archive Team Leader
BioImage Archive
EditResearch interests
EMBL-EBI’s imaging databases, the BioImage Archive and EMPIAR (Electron Microscopy Public Image Archive) are used by tens of thousands of scientists, who provide and access life sciences images. They have strong interactions with both EMBL’s world leading imaging facilities, as well as the worldwide BioImaging community. This gives us the ideal position to develop image data conversion, storage, compression, visualisation and analysis technologies. All our technology development is primarily applied to improve our service offering to imaging scientists, image analysts and researchers in the biological sciences. Automation of FAIR data management, particularly through AI processes (e.g. LLM agents, MCP servers), and application to multimodal data (e.g. spatial transcriptomic) is a key strategic goal for our service development.

Julia Mahamid
Head of Molecular Systems Biology
Molecular Systems Biology Unit
EditResearch interests
In-cell structural biology attained via cryo-electron tomography (cryo-ET) of intact cells is currently at very exciting stage; as the highest resolving technique to image the unperturbed cellular machinery in three dimensions (3D), cryo-ET produces data that can be quantitatively analyzed to derive mechanistic models of heterogeneous macromolecular assemblies within their functional context. With exceptional access to state-of-the-art infrastructure at EMBL, scientific and industry collaborations, we address experimental and computational bottlenecks in our workflows, pushing the boundaries of possible applications and increasing their robustness to make our methods more widely available together with the service teams at the EMBL Imaging Centre. The increasing users demand for these technologies necessitates developments and automation, in particular in advanced cryo-plasmaFIB volume imaging and lift-out lamella preparations for vitrified multicellular specimens, assisted by 3D correlative image registration. The increasing data volumes that are already being generated demand robust solutions for data management and flow to the public data repositories at EMBL-EBI (EMDB, EMPIAR and BioImage Archive).

Gergely Papp
Team Leader
EditResearch interests
The Papp Team at EMBL Grenoble develops scientific instrumentation for macromolecular X-ray Crystallography and Cryo-Electron Microscopy applications. One of our most recent and significant development projects is the EasyGrid technology, a system designed to fully automate sample preparation and quality control for cryo-imaging experiments. Pilot measurements demonstrated that the EasyGrid system is suitable for preparing samples for Single Particle Analysis experiments and showed the superiority of EasyGrid-prepared samples in terms of ice quality for Cryo-Electron Tomography and X-ray Nano Imaging techniques, compared to traditional plunge freezing equipment. Based on these solid technical developments, the team is now working on creating software tools to fully automate the Cryo-EM/ET sample preparation workflow, including an AI-based system for preparation parameter optimization using an on-the-fly sample quality control method.

Robert Prevedel
Group Leader and Senior Scientist
EditResearch interests
We are developing advanced optical imaging methods that are based on multi-photon microscopy, active wave-front shaping, photo-acoustics as well as high-resolution spectroscopy. Our aim is to establish our new approaches as disruptive technologies in the life sciences and to further engineer and automate our prototypes for routine service provision.

Sinem K. Saka
Group Leader
EditResearch interests
We are at the forefront of developing cutting-edge technologies that extend the boundaries of spatial biology research. Our focus is on understanding how the molecular and spatial compositions of cells are linked to their phenotypes. To achieve this, we have established state-of-the-art methods for imaging-driven spatial omics (multiplexed protein / RNA detection, imaging-base ex situ omics analyses) and have automated workflows that seamlessly integrate hardware and software for adaptive feedback microscopy, in collaboration with ALMF. Additionally, we develop and disseminate detection assays (and the associated reagent chemistries) that enable multiplexed imaging, spatial barcoding and spatial omics methods. Experimental: We are interested in spatial biology and address it with super-resolution, multiplexed imaging and spatial omics technologies that we have developed using DNA nanotechnology. We aim to push custom spatial omics approaches with 3D implementation or non-model organisms, which will be unique and very useful to many groups at EMBL and beyond to investigate the heterogeneous ecosystem of different tissues, including reproductive/developmental biology, planetary biology, microbiome and exposome studies in human tissues and disease models with a wide range of sample types (organs-on-a-chip, thick tissue slices, whole embryos or organisms, organoids, organs, xenografts). We have multiple collaborations with academic groups and industry where we perform generalized application or custom adaptation of our methods, and hence have many use cases for the service provision. Computational / data-oriented: Additionally, we are building an expansive collection of multidimensional spatial data, which is particularly well-suited for novel AI and machine learning-based analyses. The analyses results can similarly drive generation of new targets sets and development of new assays, and we would have the capacity to embed method development in this feedback system between analyses, experimental design and data generation. The datasets we generate and work with represent next generation data types and modalities that are increasingly being generated and are anticipated to be represent popular future use cases for a wide range of data-related services. Our group is dedicated to exploring the potential of advanced data analysis methods on next-generation spatial data, characterized by its rich and complex feature space. This involves both the technology development aspect, where we innovate new analytical approaches, and the support of essential infrastructure, including data formats, metadata schemas, and FAIR data management practices. These elements are vital for both research and service provision.

Christian Tischer
Team Leader
Data Science Centre
EditResearch interests
Volume electron microscopy (vEM) is a fast-evolving imaging field that aims at acquiring ultrastructural details of cells, small model organisms, and tissues in three dimensions. vEM generally produces large datasets that can reach multiple TB of data, for which extracting valuable morphometric information by image segmentation is still a challenge. For targeted studies, which restrict measurement to selected regions of interest, other imaging modalities (light microscopy (LM) or X-ray microscopy (XRM)) are used to build volumetric maps of the samples in which sub-regions of interest are defined. Due to the size and complexity of such multi-modal image datasets it is challenging to efficiently (ideally automatically) extract biophysical information from these information-rich datasets. To tackle this challenge, the Bioimage Analysis Support Team and Electron Microscopy Core Facility from EMBL Heidelberg as well as the VIB Bio Imaging Core from KU Leuven are teaming up to develop and deploy re-usable computational workflows for the automated management and (AI-based) analysis of such correlative volume EM datasets.

Andrew McCarthy
Team Leader
EditResearch interests
The McCarthy Synchrotron technology team at EMBL Grenoble operates a suite of modern structural biology beamlines with the ESRF. We continually develop new methods and automation routines for macromolecular crystallography and small angle X-ray scattering experiments. We have also started to develop biological X-ray imaging techniques in collaboration with the Papp team and our ESRF colleagues. In the context of this we are interested in receiving proposals related to either: 1) the further development of advanced in situ serial room temperature crystallography techniques and 2) the development of biological X-ray imaging applications at synchrotrons.

Timo Zimmermann
Team Leader
EditResearch interests
The team is working on a range of advanced imaging methods in the field of light microscopy (super-resolution microscopy, lightsheet imaging) and correlative light and electron microscopy (cryo-fluorescence). A strong focus of the team lies on methods that can be incorporated into service workflows in the EMBL Imaging Centre. Potential future development interests also lie in the area of multiplexed/high throughput imaging using advanced lightsheet methods and the associated data management and processing.
Structural biology and Mechanical engineering
The groups listed here are working on development of technologies in automation, robotics and/or microfluidics.

Alex Bateman
Senior Team Leader – Protein Sequence Resources
EditResearch interests
The sequence families team develops the InterPro, Pfam, Rfam and RNAcentral databases. This provides and excellent environment to learn and help develop cutting edge information resources. At present we are rapidly adopting AI technologies to assist in coding and curation. There are many opportunities and we are increasing our ambitions to provide detailed and accurate biological information more rapidly to the community. Projects could focus on using AI to improve data, or the software infrastructure for any of the data resources we provide.

Maria Bernabeu Aznar
Group Leader and Co-chair of Infection Biology Transversal Theme
EditResearch interests
Our group works to understand how vascular dysfunction contributes to infectious disease pathogenesis, by building and applying physiologically relevant 3D microvessel models. We focus on reconstructing human microvascular networks on chip to study host–pathogen interactions in a controlled, tunable environment. A major goal is to recapitulate key features of tissue-specific vasculature and use these platforms to interrogate how infections alter endothelial function, barrier properties, and tissue communication. Our lab has a strong track record in developing and integrating advanced bioengineering approaches — including microfluidic systems (mBio, Nature) and 3D tissue patterning (Science Advances)— to model human brain and liver microvasculature. Our current project focuses on engineering a liver-on-chip model to study how Plasmodium blood-stage parasites interact with the hepatic vasculature. In this call, we are particularly interested in candidates with backgrounds in bioengineering and expertise in microfluidics, organ-on-chip systems development, or mechanical engineering. Any of these three skills are essential for advancing our platform technologies and enabling their broader application across infectious disease models. That said, we are always keen to hear from creative minds who bring fresh perspectives and strong problem-solving abilities, even if their background doesn’t exactly match the profile listed here.

Alvaro Hernan Crevenna Escobar
Head of Light Imaging Facility
EditResearch interests
A biology/physics group with expertise in microscopy, histology and image analysis for two purposes: a, to further develop imaging technology; and b, to establish complex platforms such as spatial transcriptomics, tissue profiling and correlative X-ray imaging/super-resolution microscopy. We aim at bringing these services for the wider European research community through academic or industry collaborations.

Maria Marta Garcia Alai
Head of EMBL SPC Facility
EditResearch interests
Development of Software for X-ray Tomography of Sediment Samples We use X-ray tomography to explore and identify microbial ecosystems within sediment samples. The ARISE Fellow will contribute to data analysis, focusing on the visual identification and segmentation of various organisms such as diatoms and cyanobacteria. Our goal is to integrate machine learning models for automated extraction and classification, streamlining the analysis process and improving accuracy. Development of Software for Molecular Biophysics Our team is developing eSPC, an online data analysis platform for molecular biophysics (https://spc.embl-hamburg.de/). Each module follows a standardized workflow: data loading, parameter input, model selection and fitting, followed by graph production and data export. We aim to integrate AI-driven decision-making to enhance quality control, optimize model selection, and suggest follow-up experiments—streamlining analysis and reducing reliance on human expertise. This would not only improve data reliability but also broaden accessibility to non-specialists.

Matthew Hartley
BioImage Archive Team Leader
EditResearch interests
EMBL-EBI’s imaging databases, the BioImage Archive and EMPIAR (Electron Microscopy Public Image Archive) are used by tens of thousands of scientists, who provide and access life sciences images. They have strong interactions with both EMBL’s world leading imaging facilities, as well as the worldwide BioImaging community. This gives us the ideal position to develop image data conversion, storage, compression, visualisation and analysis technologies. All our technology development is primarily applied to improve our service offering to imaging scientists, image analysts and researchers in the biological sciences. Automation of FAIR data management, particularly through AI processes (e.g. LLM agents, MCP servers), and application to multimodal data (e.g. spatial transcriptomic) is a key strategic goal for our service development.

Alexandra Koumoutsi
Head of Microbial Automation and Culturomics Core Facility
EditResearch interests
The Microbial Automation and Culturomics Core Facility will provide access and training to cutting-edge equipment and knowhow related to automation for microbial work, with an initial focus on the human gut microbiome. Moreover, the facility will enable the deconvolution of microbial complex communities (isolation of most/ all species within a community). Using robotic systems inside/ outside of controlled environmental chambers, researchers will be able to construct and characterize genome-wide mutant libraries, assemble strain collections or communities and perform large-scale screens (from phenotyping to compound screens). The facility aims to create and provide genetic tools and resources for non-model species and microbial communities and establish analysis pipelines for genomic to high-throughput phenotypic data.

Julia Mahamid
Head of Molecular Systems Biology
EditResearch interests
In-cell structural biology attained via cryo-electron tomography (cryo-ET) of intact cells is currently at very exciting stage; as the highest resolving technique to image the unperturbed cellular machinery in three dimensions (3D), cryo-ET produces data that can be quantitatively analyzed to derive mechanistic models of heterogeneous macromolecular assemblies within their functional context. With exceptional access to state-of-the-art infrastructure at EMBL, scientific and industry collaborations, we address experimental and computational bottlenecks in our workflows, pushing the boundaries of possible applications and increasing their robustness to make our methods more widely available together with the service teams at the EMBL Imaging Centre. The increasing users demand for these technologies necessitates developments and automation, in particular in advanced cryo-plasmaFIB volume imaging and lift-out lamella preparations for vitrified multicellular specimens, assisted by 3D correlative image registration. The increasing data volumes that are already being generated demand robust solutions for data management and flow to the public data repositories at EMBL-EBI (EMDB, EMPIAR and BioImage Archive).

José Márquez
Head of Crystallisation Facility
EditResearch interests
Our Team has pioneered the development of Online Crystallography; fully automated protein-to structure pipelines integrating crystallization, synchrotron data collection and crystallographic data analysis into continuous workflows operated via the web. These pipelines are currently used by hundreds of scientists worldwide and are based on the CrystalDirect technologies and CRIMS software, which we have contributed to develop. Recently, we have implemented a fully automated pipeline for ligand and fragment screening to support structure guided drug design. EMBL Grenoble is co-located with the European Synchrotron Radiation Facility (ESRF) in Grenoble, which produces some of the world’s most brilliant X-ray beams worldwide. EMBL and ESRF jointly operate six crystallography beamlines one of which is the fully automated MASSIF-1 whose operation is highly integrated with the EMBL’s HTX Lab. Our interdisciplinary team offers opportunities for scientists, engineers and software developers to work in one of the leading infrastructures for structural biology within the areas of protein crystallography, structure-based drug design, automation, and large-scale scientific data management and analysis. Currently, we are particularly interested in profiles either in structural biology or computer science orientated towards one or several of the following areas: X-ray-based fragment screening, structure guided drug design, AI-based and advanced computational approaches for hit-to-lead optimisation, medicinal chemistry.

Gergely Papp
Team Leader
EditResearch interests
The Papp Team at EMBL Grenoble develops scientific instrumentation for macromolecular X-ray Crystallography and Cryo-Electron Microscopy applications. One of our most recent and significant development projects is the EasyGrid technology, a system designed to fully automate sample preparation and quality control for cryo-imaging experiments. Pilot measurements demonstrated that the EasyGrid system is suitable for preparing samples for Single Particle Analysis experiments and showed the superiority of EasyGrid-prepared samples in terms of ice quality for Cryo-Electron Tomography and X-ray Nano Imaging techniques, compared to traditional plunge freezing equipment. Based on these solid technical developments, the team is now working on creating software tools to fully automate the Cryo-EM/ET sample preparation workflow, including an AI-based system for preparation parameter optimization using an on-the-fly sample quality control method.

Robert Prevedel
Group Leader and Senior Scientist
EditResearch interests
We are developing advanced optical imaging methods that are based on multi-photon microscopy, active wave-front shaping, photo-acoustics as well as high-resolution spectroscopy. Our aim is to establish our new approaches as disruptive technologies in the life sciences and to further engineer and automate our prototypes for routine service provision.

Sinem K. Saka
Group Leader
EditResearch interests
We are at the forefront of developing cutting-edge technologies that extend the boundaries of spatial biology research. Our focus is on understanding how the molecular and spatial compositions of cells are linked to their phenotypes. To achieve this, we have established state-of-the-art methods for imaging-driven spatial omics (multiplexed protein / RNA detection, imaging-base ex situ omics analyses) and have automated workflows that seamlessly integrate hardware and software for adaptive feedback microscopy, in collaboration with ALMF. Additionally, we develop and disseminate detection assays (and the associated reagent chemistries) that enable multiplexed imaging, spatial barcoding and spatial omics methods. Experimental: We are interested in spatial biology and address it with super-resolution, multiplexed imaging and spatial omics technologies that we have developed using DNA nanotechnology. We aim to push custom spatial omics approaches with 3D implementation or non-model organisms, which will be unique and very useful to many groups at EMBL and beyond to investigate the heterogeneous ecosystem of different tissues, including reproductive/developmental biology, planetary biology, microbiome and exposome studies in human tissues and disease models with a wide range of sample types (organs-on-a-chip, thick tissue slices, whole embryos or organisms, organoids, organs, xenografts). We have multiple collaborations with academic groups and industry where we perform generalized application or custom adaptation of our methods, and hence have many use cases for the service provision. Computational / data-oriented: Additionally, we are building an expansive collection of multidimensional spatial data, which is particularly well-suited for novel AI and machine learning-based analyses. The analyses results can similarly drive generation of new targets sets and development of new assays, and we would have the capacity to embed method development in this feedback system between analyses, experimental design and data generation. The datasets we generate and work with represent next generation data types and modalities that are increasingly being generated and are anticipated to be represent popular future use cases for a wide range of data-related services. Our group is dedicated to exploring the potential of advanced data analysis methods on next-generation spatial data, characterized by its rich and complex feature space. This involves both the technology development aspect, where we innovate new analytical approaches, and the support of essential infrastructure, including data formats, metadata schemas, and FAIR data management practices. These elements are vital for both research and service provision.

Nassos Typas
Head of Molecular Systems Biology
EditResearch interests
The Typas lab develops high-throughput approaches (genetics and biochemical) to systematically study microbial physiology, gene functions and interactions. This involves coupling genetic and biochemical-based approaches to quantitative multi-parametric phenotyping (from fitness-based to microscopy and omics readouts) to investigate gene function and organization in model, and recently non-model bacteria. Robotic automation (liquid handling, colony picking robots, stackers-readers) and large data analysis are integral parts of this process. In the microbiome area, we use our abilities for high-throughput cultivation, characterisation and genetic manipulation of microbial communities. Together with 15 other labs at EMBL (and alumni) and as part of the Microbial Ecosystems Transversal Theme, we are working towards generating the tools, resources and functional information required for establishing from scratch new model species for the human gut microbiome. Knowledge and approaches that we develop move to the different facilities at EMBL, including the MAC, PEP and proteomics core facilities – all possible collaborators here. As part of this call, opportunities include: a) developing pipelines for systematically map protein-ligand interactions in gut microbial species and communities; b) developing genome-wide genetic tools for understudied but important gut bacteria; and c) establishing model communities for the human gut. Within this realm, multiple opportunities exist for self-designed projects in areas at the interface of microbiome and systems biology, such as developing pipelines to systematically map protein-ligand interactions; developing genome-wide genetic tools for understudied but important bacteria; and establishing model communities for microbiomes.

Andrew McCarthy
Team Leader
EditResearch interests
The McCarthy Synchrotron technology team at EMBL Grenoble operates a suite of modern structural biology beamlines with the ESRF. We continually develop new methods and automation routines for macromolecular crystallography and small angle X-ray scattering experiments. We have also started to develop biological X-ray imaging techniques in collaboration with the Papp team and our ESRF colleagues. In the context of this particular ARISE2 call we are potentially looking for two fellows interested in either: 1) the further development of advanced in situ serial room temperature crystallography techniques and 2) the development of biological X-ray imaging applications at synchrotrons.

Kyle Morris
EMDB Team Leader
Electron Microscopy Data Bank
EditBiotechnology
The groups listed here are working on development of technologies in (bio)chemical engineering and genetic engineering

Maria Bernabeu Aznar
Group Leader and Co-chair of Infection Biology Transversal Theme
EditResearch interests
Our group works to understand how vascular dysfunction contributes to infectious disease pathogenesis, by building and applying physiologically relevant 3D microvessel models. We focus on reconstructing human microvascular networks on chip to study host–pathogen interactions in a controlled, tunable environment. A major goal is to recapitulate key features of tissue-specific vasculature and use these platforms to interrogate how infections alter endothelial function, barrier properties, and tissue communication. Our lab has a strong track record in developing and integrating advanced bioengineering approaches — including microfluidic systems (mBio, Nature) and 3D tissue patterning (Science Advances)— to model human brain and liver microvasculature. Our current project focuses on engineering a liver-on-chip model to study how Plasmodium blood-stage parasites interact with the hepatic vasculature. In this call, we are particularly interested in candidates with backgrounds in bioengineering and expertise in microfluidics, organ-on-chip systems development, or mechanical engineering. Any of these three skills are essential for advancing our platform technologies and enabling their broader application across infectious disease models. That said, we are always keen to hear from creative minds who bring fresh perspectives and strong problem-solving abilities, even if their background doesn’t exactly match the profile listed here.

Michael Dorrity
Group Leader
EditResearch interests
The Dorrity lab develops new technologies to for statistical developmental genetics, focusing on (1) new molecular approaches for ultra-scalable multi-omic single cell profiling and (2) new computational methods and software to manage and analyze the multi-scale data resulting from these experiments. We are also collaborating with the “Frontier” Organism Center to generate systematic spatiotemporal perturbation datasets to learn how the environment affects cellular and organismal function. We have opportunities for potential ARISE fellows at each level.

Alexandra Koumoutsi
Head of Microbial Automation and Culturomics Core Facility
EditResearch interests
The Microbial Automation and Culturomics Core Facility will provide access and training to cutting-edge equipment and knowhow related to automation for microbial work, with an initial focus on the human gut microbiome. Moreover, the facility will enable the deconvolution of microbial complex communities (isolation of most/ all species within a community). Using robotic systems inside/ outside of controlled environmental chambers, researchers will be able to construct and characterize genome-wide mutant libraries, assemble strain collections or communities and perform large-scale screens (from phenotyping to compound screens). The facility aims to create and provide genetic tools and resources for non-model species and microbial communities and establish analysis pipelines for genomic to high-throughput phenotypic data.

José Márquez
Head of Crystallisation Facility
EditResearch interests
Our Team has pioneered the development of Online Crystallography; fully automated protein-to structure pipelines integrating crystallization, synchrotron data collection and crystallographic data analysis into continuous workflows operated via the web. These pipelines are currently used by hundreds of scientists worldwide and are based on the CrystalDirect technologies and CRIMS software, which we have contributed to develop. Recently, we have implemented a fully automated pipeline for ligand and fragment screening to support structure guided drug design. EMBL Grenoble is co-located with the European Synchrotron Radiation Facility (ESRF) in Grenoble, which produces some of the world’s most brilliant X-ray beams worldwide. EMBL and ESRF jointly operate six crystallography beamlines one of which is the fully automated MASSIF-1 whose operation is highly integrated with the EMBL’s HTX Lab. Our interdisciplinary team offers opportunities for scientists, engineers and software developers to work in one of the leading infrastructures for structural biology within the areas of protein crystallography, structure-based drug design, automation, and large-scale scientific data management and analysis. Currently, we are particularly interested in profiles either in structural biology or computer science orientated towards one or several of the following areas: X-ray-based fragment screening, structure guided drug design, AI-based and advanced computational approaches for hit-to-lead optimisation, medicinal chemistry.

Gerald Pfister
Head of Flow Cytometry Facility
EditResearch interests
A primary technology development focus at the EMBL Rome FCF is advanced flow cytometry and cell sorting. These tools are crucial for accurate single-cell data acquisition and for purifying cells for diverse downstream applications like RNA/DNA analysis and single-cell sequencing. Current projects include developing & optimizing flow cytometry protocols for a number of different samples & cell types. Bulk- and single-cell sorting assays of various different sample- and cell types are routinely designed and performed. Recently, we started to set up assays for soluble protein analysis by flow cytometry, and are exploring novel imaging-enabled flow cytometry assays. Currently, two high-end cell sorters (BD FACSAria Fusion with BSL-2 cabinet & BD FACSMelody), an Attune NxT flow cytometer with autosampler, and a 10X Genomics Chromium controller are available in the facility For ARISE2, the FCF, as the host lab and main technology development partner in advanced flow cytometry and cell sorting techniques, is seeking collaborative project proposals. These projects will leverage our cutting-edge capabilities with robust bioinformatics approaches. We are particularly interested in proposals that align with the biological research focus of our collaborator, the Rompani lab. This includes generating and analyzing complex datasets to explore molecular underpinnings of biological phenomena, such as those related to age-related changes or cellular profiling linked to higher-level phenotypic data. A strong bioinformatics component, centered on developing novel analytical tools and integrated interpretation pipelines, is essential for these projects. This integrated approach, emphasizing the creation of new analytical methods rather than mere application, will offer comprehensive data lifecycle experience, fostering technology creation.

Sinem K. Saka
Group Leader
EditResearch interests
We are at the forefront of developing cutting-edge technologies that extend the boundaries of spatial biology research. Our focus is on understanding how the molecular and spatial compositions of cells are linked to their phenotypes. To achieve this, we have established state-of-the-art methods for imaging-driven spatial omics (multiplexed protein / RNA detection, imaging-base ex situ omics analyses) and have automated workflows that seamlessly integrate hardware and software for adaptive feedback microscopy, in collaboration with ALMF. Additionally, we develop and disseminate detection assays (and the associated reagent chemistries) that enable multiplexed imaging, spatial barcoding and spatial omics methods. Experimental: We are interested in spatial biology and address it with super-resolution, multiplexed imaging and spatial omics technologies that we have developed using DNA nanotechnology. We aim to push custom spatial omics approaches with 3D implementation or non-model organisms, which will be unique and very useful to many groups at EMBL and beyond to investigate the heterogeneous ecosystem of different tissues, including reproductive/developmental biology, planetary biology, microbiome and exposome studies in human tissues and disease models with a wide range of sample types (organs-on-a-chip, thick tissue slices, whole embryos or organisms, organoids, organs, xenografts). We have multiple collaborations with academic groups and industry where we perform generalized application or custom adaptation of our methods, and hence have many use cases for the service provision. Computational / data-oriented: Additionally, we are building an expansive collection of multidimensional spatial data, which is particularly well-suited for novel AI and machine learning-based analyses. The analyses results can similarly drive generation of new targets sets and development of new assays, and we would have the capacity to embed method development in this feedback system between analyses, experimental design and data generation. The datasets we generate and work with represent next generation data types and modalities that are increasingly being generated and are anticipated to be represent popular future use cases for a wide range of data-related services. Our group is dedicated to exploring the potential of advanced data analysis methods on next-generation spatial data, characterized by its rich and complex feature space. This involves both the technology development aspect, where we innovate new analytical approaches, and the support of essential infrastructure, including data formats, metadata schemas, and FAIR data management practices. These elements are vital for both research and service provision.

Nassos Typas
Head of Molecular Systems Biology
EditResearch interests
The Typas lab develops high-throughput approaches (genetics and biochemical) to systematically study microbial physiology, gene functions and interactions. This involves coupling genetic and biochemical-based approaches to quantitative multi-parametric phenotyping (from fitness-based to microscopy and omics readouts) to investigate gene function and organization in model, and recently non-model bacteria. Robotic automation (liquid handling, colony picking robots, stackers-readers) and large data analysis are integral parts of this process. In the microbiome area, we use our abilities for high-throughput cultivation, characterisation and genetic manipulation of microbial communities. Together with 15 other labs at EMBL (and alumni) and as part of the Microbial Ecosystems Transversal Theme, we are working towards generating the tools, resources and functional information required for establishing from scratch new model species for the human gut microbiome. Knowledge and approaches that we develop move to the different facilities at EMBL, including the MAC, PEP and proteomics core facilities – all possible collaborators here. Within this realm, multiple opportunities exist for self-designed projects in areas at the interface of microbiome and systems biology, such as developing pipelines to systematically map protein-ligand interactions; developing genome-wide genetic tools for understudied but important bacteria; and establishing model communities for microbiomes.
Partner organisations
A number of partner organisations are participating in the ARISE2 programme.
Some are open to full collaborations where they support fellows during the 3-year research projects offering additional expertise and technologies that may not be available at EMBL.





Many partners offer secondment opportunities (2-6 months long secondments or 1-2 week short visits) where they collaborate with fellows on a part of the research project or provide job shadowing opportunities. If the partner is collaborating on the full 3-year research project, fellows may spend up to 11 months on secondment with them.

ARINAX







Luxendo Bruker





VBCF


Zeiss
The programme also has an implementing partner, RIcapacity, who is involved in the development of partnerships and the training programme in professional management skills for research infrastructure scientists. Associated partner, BII, will also contribute specifically to the training programme. Based on topics/themes, the following associated partners will also contribute their expertise to the training programme: ARINAX, CEITEC, Cellzome/GSK, CRG, ESRF, Leica, Luxendo Bruker, SciLifeLab, UGA, VIB

RIcapacity

BII

Duration of fellowship: 3 years
Number of fellowships: 17
Dates: 2025 Call – Closed
Call opening: 30.06.2025
Call closing: 30.09.2025
Interviews: Week of December 1st, 2025
Fellowship Start: 01.01.2026 (or within next 4 months)
A detailed call schedule is available in the guide to applicants.
Contact: arise2@embl.org

