Michael Dorrity
Group Leader
michael.dorrity [at] embl.de
ORCID: 0000-0002-7691-6504
EditEnvironmental response at the single-cell level

Group Leader
michael.dorrity [at] embl.de
ORCID: 0000-0002-7691-6504
EditEffects of environmental stress are variable, with some individuals more sensitive than others. This variability obscures the influence of the environment during development, limits our understanding of genotype–phenotype relationships, and leaves us unable to predict how a changing climate will shape populations and ecologies globally.
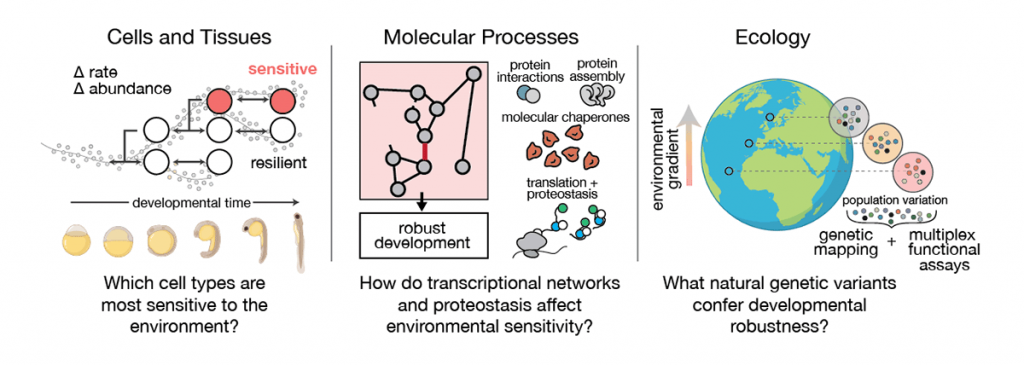
To better understand how environmentally induced variability propagates across scales, the lab uses single-cell genomics as a phenotyping tool to capture cell- and tissue-level processes, along with molecular phenotypes. We conduct this multiscale phenotyping in individual zebrafish embryos, a model for aquatic vertebrate development. Using computational and statistical approaches to partition variability at each level (molecular, cellular, and organismal), we can define sources of environmental responsiveness and uncover potential mechanisms of environmental adaptation. We validate and explore these mechanisms experimentally by targeting key factors using dominant negative protein fragments, gene editing, and pharmacological inhibitors.
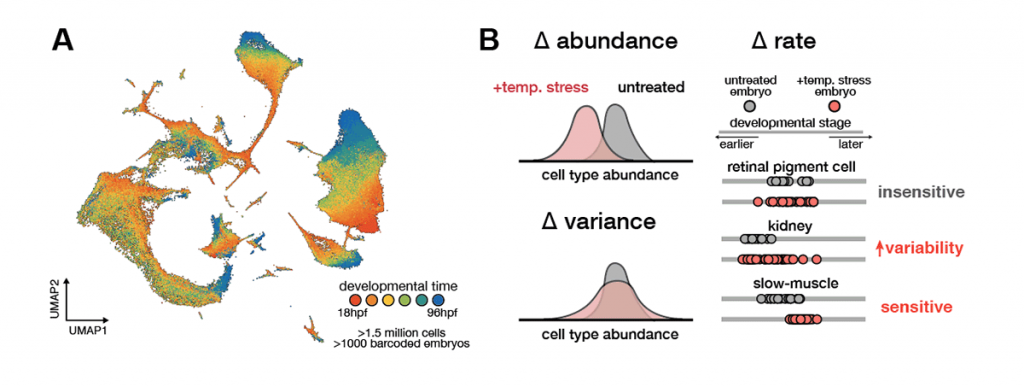
Our previous work has shown that raising zebrafish embryos in elevated water temperatures has profound effects on the programme of development: abundances of cell types are altered in the embryo, specific cell types show increased variability across individual embryos, and temperature-dependent changes to developmental rate are introduced non-uniformly across cell types. These findings reveal how a broad stressor like temperature nevertheless induces cell type-specific effects, which underlie variable whole-animal phenotypes.
At the molecular level, cell type-specific stress sensitivity can be linked to components of proteostasis, or the maintenance of proper folding, synthesis, and degradation of proteins in cells. To investigate the role of proteostasis in environmentally induced variability at the molecular level, we have also developed multiplex assays of protein function. Leveraging high-throughput sequencing to characterise protein variants in yeast, we can identify dominant negative protein fragments that inhibit a target of interest, and we can determine functional consequences of hundreds of thousands of missense mutations for a given protein. We can use these high-throughput screens to identify variants that disrupt or alter function of proteins or pathways involved in environmental sensing.
We will build on these experimental and computational tools to predict how a changing climate will influence animal development across scales. Activities will include: