Novel algorithm able to detect mutations in single-cell sequencing data sets
SComatic enables researchers to link genotype to phenotype using only single-cell profiling data
SCIENCE & TECHNOLOGY2023
research-highlightssciencescience-technology
Showing results out of

SComatic enables researchers to link genotype to phenotype using only single-cell profiling data
SCIENCE & TECHNOLOGY2023
research-highlightssciencescience-technology

Two former EMBL scientists have been recognised for their outstanding contributions to the fields of science communication and multiple sequence alignment research, respectively.
2023
alumni

EMBL Hamburg’s Grzegorz Chojnowski from the Wilmanns Group developed software called findMySequence, which identifies proteins’ amino-acid sequences based on electron cryo-microscopy and X-ray crystallography data. It’s useful for identifying unknown proteins in samples from natural sources.
SCIENCE & TECHNOLOGY2022
sciencescience-technology
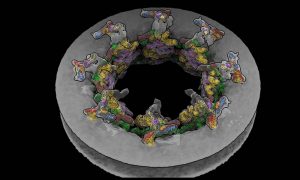
EMBL Hamburg’s Kosinski Group, the Beck Laboratory at the Max Planck Institute of Biophysics, and colleagues at EMBL Heidelberg recorded the nuclear pore complex contracting in living cells. They visualised the movement with an unprecedented level of detail with help of new software called…
SCIENCE & TECHNOLOGY2021
sciencescience-technology
The Chan Zuckerberg Initiative has recognised four EMBL researchers with their most recent awards, showing how tech trailblazers are integral to advancing science and medicine.
EMBL ANNOUNCEMENTSLAB MATTERS2020
embl-announcementslab-matters
Coin toss inspires CorMap: a new statistical test that sidesteps need for error estimation.
SCIENCE & TECHNOLOGY2015
sciencescience-technology
In the early days of X-ray crystallography obtaining a three-dimensional model of a protein required wire models, screws, bolts and years of tedious calculations by hand. Today macromolecular models are built by computers – thanks to sophisticated software and in particular a package called…
LAB MATTERSSCIENCE & TECHNOLOGY2006
lab-mattersscience-technology
EMBOSS, the European Molecular Biology Open Software Suite, has received a vital funding boost from the UK Biotechnology and Biological Sciences Research Council (BBSRC) that will guarantee its continued maintenance under an open source license for the next three years. This ends two years of…
CONNECTIONSLAB MATTERS2006
connectionslab-matters
No results found