
Reconstructing 3D shapes from unmeasurable data
Scientists at EMBL Hamburg determine shapes of intermediate states in dynamic biological systems
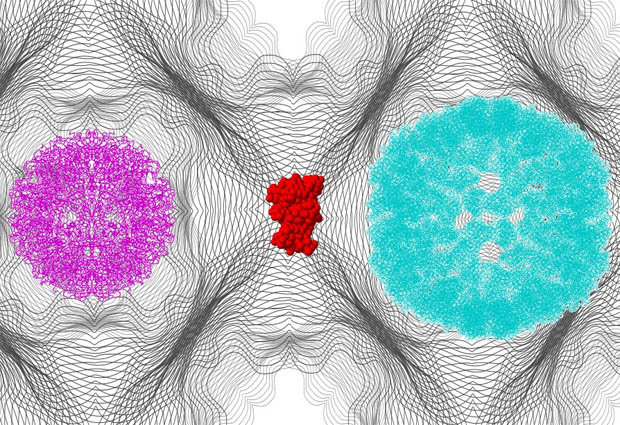
A big challenge in modern structural biology is the characterisation of dynamic biological systems. For example, the formation of amyloid fibrils: aggregates of proteins that play a major role in neurodegenerative diseases like Alzheimer’s. The intermediates of such processes are hard to study, because they always co-exist with initial and final states.
Dmitri Svergun, group leader at EMBL Hamburg, succeeded in reconstructing the 3D shape of such unmeasurable intermediate macromolecular states, together with Petr Konarev from the Crystallography and Photonics Research Center in Moscow. The Journal of the International Union of Crystallography (IUCrJ) publishes their new method on May 29.
Svergun and Konarev reconstructed the shapes directly from multiple small-angle X-ray scattering (SAXS) datasets taken on an evolving system. The new approach will be useful for interpreting a broad range of experiments on dynamic biological systems and intermediate states, such as protein folding and unfolding, or viral self-assembly during replication.
Video explaining small-angle X-ray scattering. CREDIT: WeNMR, Spronk 3D studio & EMBL Svergun group.


