
Read the latest Issue
New study demonstrates how monitoring all cellular proteins over time and space can improve our understanding of host–pathogen interactions
When humans get infected by pathogenic bacteria, the body’s immune system tries to eliminate the intruders. One way of doing this is by launching an inflammatory response – a cascade of events that includes the expression of protective proteins, the activation of immune cells, and a process of controlled cell death when infected cells can’t be saved.
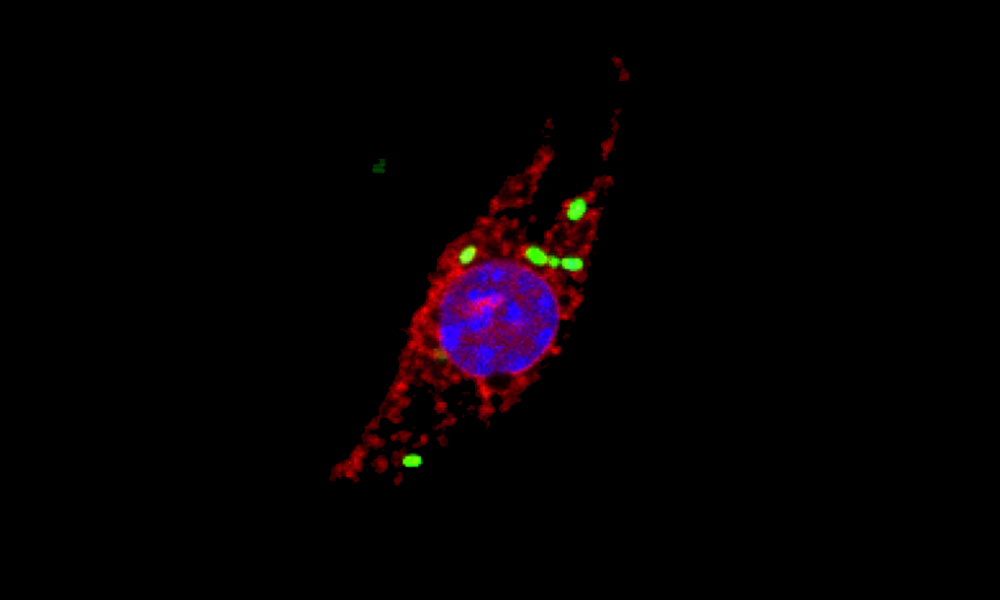
Scientists including members of EMBL’s Typas group, members of the group of EMBL alumnus Jeroen Krijgsveld at the German Cancer Research Center (DKFZ) in Heidelberg, and other collaborators have investigated how immune cells called macrophages respond to infection by the intracellular pathogen Salmonella enterica. The scientists applied a method recently developed in their labs to enrich, identify, and quantify all newly produced proteins from Salmonella-infected macrophages. They marked newly produced proteins with a specific chemical label and identified them using a technique called mass spectrometry, which allowed them to analyse the entire set of cellular proteins. Importantly, the scientists measured protein levels in macrophages at different infection stages and across different cell compartments. Their study, which is published in Nature Microbiology, shows that monitoring the dynamic changes in protein production and targeting can reveal new insights into the mechanisms by which cells respond to pathogens.
One of the unexpected findings of the study was that a well-known family of proteins called cathepsins move to a new location when cells get infected by Salmonella. Cathepsins are proteases – proteins that break down other proteins. They’re normally kept inside small subcellular structures known as lysosomes and have previously been implicated in promoting cell death, although the mechanism or any link between the process and bacterial infection were unknown. The scientists have now discovered that Salmonella causes newly produced cathepsins to accumulate in the nuclei of infected cells. The protein-degrading activity of cathepsins in the nucleus is then required to initiate an inflammatory form of programmed cell death.
The new study shows the benefit of systematically following protein dynamics during infection, which can unravel new pathways and mechanisms the host uses to defend itself against pathogens.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive