
New perspectives on nuclear pores
EMBL researchers have published two new studies involving the nuclear pore complex
From a tiny yeast specimen to a full-grown elephant, the defining feature of organisms known as eukaryotes is that they store their genetic information in the nucleus of their cells, surrounded by a protective double membrane. Embedded into the double membrane, nuclear pore complexes (NPCs) are large assemblies of about 1000 individual proteins called nucleoporins. The function of the NPC is that of a gateway between the nucleus and the rest of the cell. NPCs therefore play a substantial role in the exchange of large molecules and in regulating gene expression.
Many research groups at EMBL have studied the NPC. In 2016, scientists in the Beck group revealed the structure of the nuclear pore’s inner ring, and, in cooperation with the Ellenberg group, described its formation across the nucleus’s double membrane. Both the Beck group and the Ries group at EMBL have recently published new studies involving the NPC, addressing very different questions. The Beck group has uncovered a previously unknown formation process for the NPC, while the Ries group describes its use as a reference standard for super-resolution microscopy.
Observing disorder
“Our study describes the assembly of the nuclear core complex,” says Bernhard Hampoelz, a visiting researcher in the Beck group and lead author of the paper. “This is a very complicated process, because a thousand proteins need to assemble in the right order, in a relatively short time. Previously, we knew about two pathways that describe how this happens in tissue culture cells.” By studying the fruit fly Drosophila, the researchers were able to describe a third, alternative assembly mechanism that occurs during oogenesis: the formation of a fertile egg cell, or oocyte.
The two previously known pathways for NPC assembly take place at the nucleus, during different stages of the cell cycle. In the oocyte, NPCs are assembled outside the nuclear compartment and stored in membrane sheets called annulate lamellae. The oocyte prepares a stockpile of NPCs that are then inherited by the embryo and inserted into the nuclear envelope during its fast cell division cycles.

Andre Schwarz, a PhD student in the Beck group, explains that the study was done by creating a transgenic fly line that expressed fluorescent proteins in its NPCs, and observing NPC assembly using light and electron microscopy. “The NPC is deeply integrated into the double membrane, so it’s basically impossible to extract and purify,” says Schwarz. “We had to do everything in vivo. That meant dissecting Drosophila ovaries, culturing them in an imaging medium and then imaging them. Working in vivo is always more difficult.”
“But also a lot cooler,” Hampoelz adds with a grin. “You get to see all these processes in a living organism, in their natural context.”
Research in the Beck group has often focused on the structure of large molecular assemblies. “In our case, we go one step back, to the assembly process itself,” says Hampoelz. “There, everything still seems to be very disordered and very dynamic. We’re interested in seeing how such a complex machine is built out of a thousand proteins. It seems that, in oocytes, this happens from a very chaotic state. How exactly the assembly is regulated and the complexes are built up piece by piece in the oocyte is something we still don’t know much about.”
A new standard
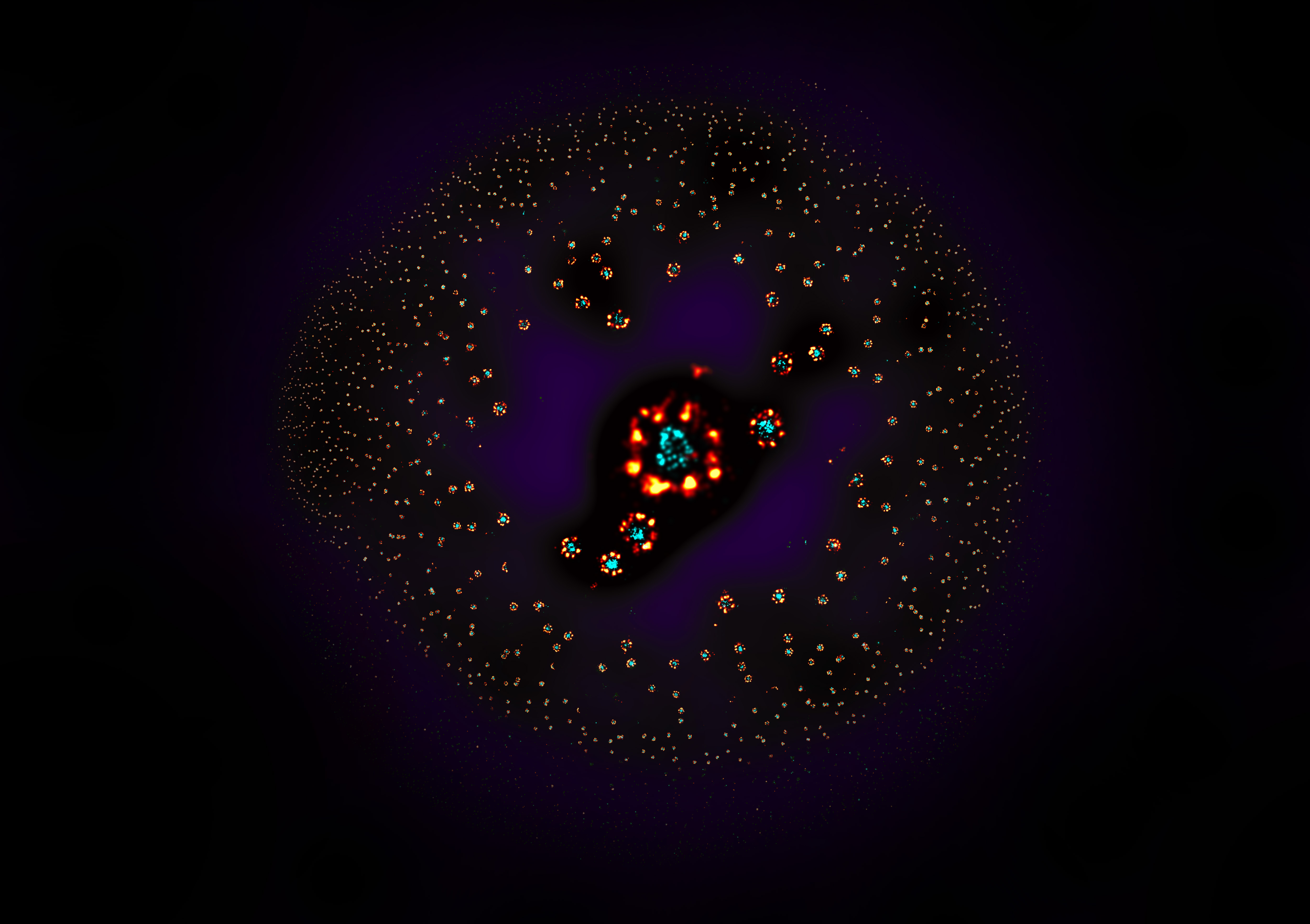
The paper from the Ries group describes the use of NPCs as a reference standard for super-resolution microscopy. This was needed to benchmark the microscopy techniques that the group develops.
“The idea to use nuclear pores as a reference standard was quite straightforward,” says group leader Jonas Ries. “The Ellenberg group was generating cell lines with labelled nucleoporins for their study of NPC biology, which happened to be perfect as a proof of concept. Once we confirmed that they worked really well, we started making cell lines with a labelled protein that was well positioned within the NPC to aid with visualisation and image analysis.”
Jervis Thevathasan, the PhD student who oversaw the project, explains its rationale. “The idea behind this was to make a tool that would help super-resolution microscopy become a quantitative method,” he says. “We needed to have an objective control over our measurements, especially for quantitative conclusions. This was something that was really missing in the field.”
Super-resolution microscopy can go to the extent of localising single molecules on a nanometre scale. This requires optimised settings for many variables, such as stability, labelling technologies, sample preparation, imaging conditions, and analysis software. The lack of a common standard makes it difficult to detect errors in these settings. Because the NPC has a very stereotypical arrangement and number of proteins, it is a suitable reference for measuring microscope performance.

The study also brought some unexpected findings. “One would think that the nuclear pore complex, like a Lego piece, should be very rigid and everything should look the same,” says Ries. “But, when we looked at it, we found a subset of nuclear pores that looked very different. I think this is something that was not really appreciated before.” Fortunately, the variation is so small that it does not interfere with using nuclear pores as a reference standard. The accuracy is very high, although not perfect. “The fact that it’s not perfect is what makes it so interesting,” Ries adds, “because it’s linked to the biology of the nuclear pore complex, which apparently is not as rigid as we thought.”
The aim of the group is that their use of NPCs as a reference standard will soon be adopted as common practice for super-resolution microscopy. “Many groups already have our cell lines and they’re also available in the cell line repository,” says Ries. “There are a number of publications coming up that use these cell lines, and it’s our hope that groups will continue using them on a regular basis.”
The study is a result of contributions from almost every member of the Ries group. This included characterisation and preparation of samples, software development, imaging, and finding ways to analyse images automatically. “An exciting aspect of biology right now is that once we know the structure of a protein or protein complex, we can think about how to reuse these geometries for other needs,” says Thevathasan. “What I really liked about this project was that we used insights from basic science to develop a tool for the broader community.”


