
Read the latest Issue
EMBL scientists create a turnover catalogue of almost 10.000 proteins from primary cells
Proteins perform countless functions in the cell, including transporting molecules, speeding up metabolic reactions and forming structural parts of the cell such as the nuclear pore complex. Protein turnover is a measure of the difference between protein synthesis and protein degradation and it is an important indicator of a cell’s activity in health and disease.
EMBL group leaders Mikhail Savitski and Martin Beck, in close collaboration with Cellzome scientists Marcus Bantscheff and Toby Mathieson, have improved the accuracy of the detection of small changes in protein turnover by developing a better algorithmic treatment of raw mass spectrometry data. As a result, the researchers have published a turnover catalogue of 9699 unique proteins in Nature Communications. The paper focuses on protein complexes and demonstrates that subunits of protein complexes have consistent turnover rates.
We wanted to study protein homeostasis, or the balanced process behind protein synthesis and degradation in primary cells extracted from blood or living tissue. Primary cells provide a better understanding of the in vivo situation than cultured cells but, unfortunately, they have a short lifespan when compared to the protein complexes we wanted to study. To overcome this problem, we developed a better algorithmic treatment of raw mass spectrometry data. The improved algorithm accurately determines very small changes in proteins, allowing us to measure the turnover of 9699 unique proteins, including very long-lived proteins, such as the Histone H1.2 protein which has a half-life of 2242 hours. For the first time, we have a view of protein turnover at a cellular scale in several primary cell types, which will be a valuable resource for the scientific community.
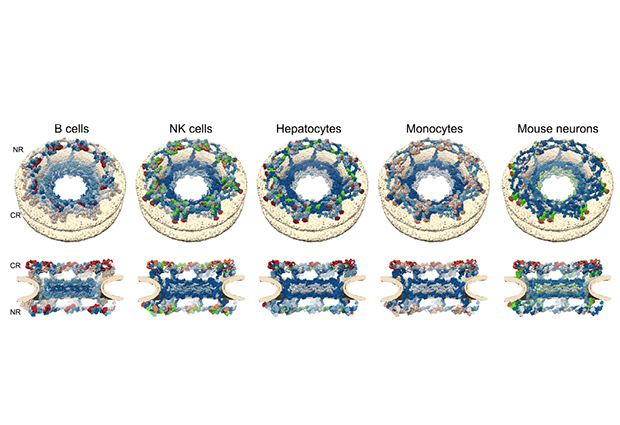
We focused our analysis on protein complexes, particularly on the nuclear pore complex, which is very big and is composed of several sub-complexes. We discovered that there are protein turnover levels that are specific to a given sub-complex. Proteins which are peripheral to the complex, that joined later in evolution, turn out to have much faster turnover than the ones that form the core structure and have been there for a longer time. Contrary to previous understanding, our data clearly suggests that there is a turnover mechanism for the nuclear pore in non-dividing cells. This is exciting because it opens new research in this direction.
Protein turnover is important for understanding cellular homeostasis. Our work delineates the tools to study the mechanisms controlling it and will help researchers study a wide range of things, such as ageing, brain function, cancer and neurodegeneration.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive