
Deep learning captures cell cycle
DeepCycle method reconstructs a complex biological phenomenon just from how cells look, opening up a novel way to better understand the cell cycle ‘clock’
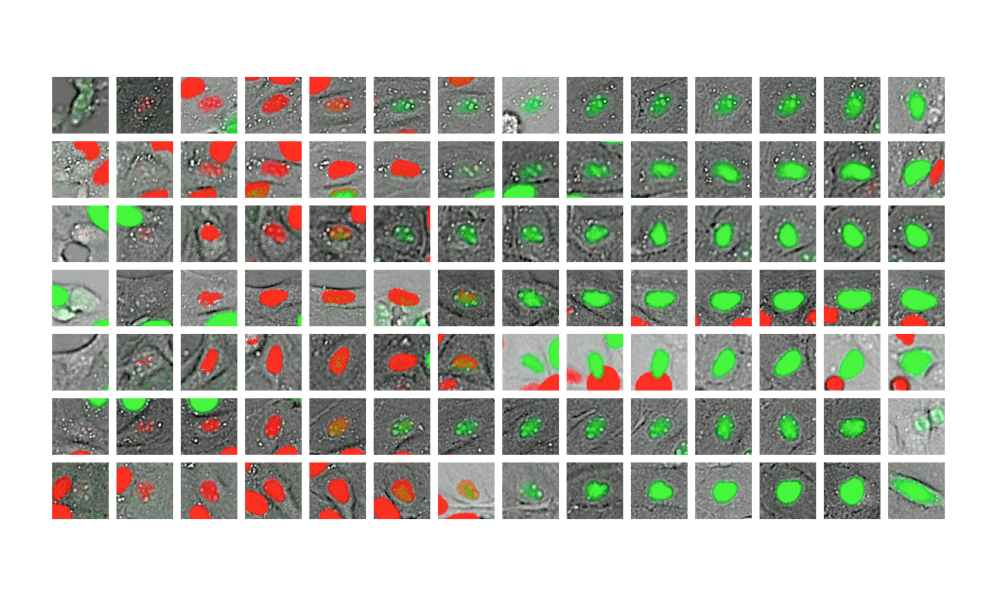
Using approximately 2.6 million microscopy images of canine kidney cells, members of an EMBL-led research group with collaborators in Estonia and Russia have built and trained a deep learning model to better understand how cells grow and divide. This research opens a novel way to identify more quickly and more easily the cell cycle ‘clock’ for every cell, which is essential to understand diseases like cancer. The methods and results of their work have now been published in the journal Molecular Systems Biology.
“It’s a pretty advanced use of deep learning,” says Theodore Alexandrov, an EMBL research team leader and paper co-author. “Rather than using any existing model, we came up with a strategy to make a deep learning model to learn about subtle differences of the cells. This ultimately led to the model being able to reconstruct the cell cycle trajectory. Intriguingly, the model learned such a complex phenomenon by itself – just from how cells look.”
Cells grow and divide in a cycle that is fairly systematic, with well-described key phases. By employing conventional microscopy on a large scale, Luca Rappez and other members of the Alexandrov team have observed cells over 33 hours, accumulating millions of images and capturing their progression through the cell cycle and cell divisions. To validate their model, they used cells with fluorescent markers that light up in different phases of the cell cycle. This enabled them to collect a multitude of images of cells at various points of their life: as they grow, when they’re on the verge of dividing, and when they actually divide and the process begins anew.
With deep learning experts from Neuromation – a small start-up in Tallinn, Estonia – and from the Steklov Mathematical Institute and the Higher School of Economics in St. Petersburg, Russia, Theodore’s group generated millions of images to train their deep learning network, called DeepCycle, to discern minute differences and similarities between cells. This was challenging, since even with the naked eye it can be hard to recognise the shapes of individual cells in these high-rate images. Following the so-called ‘transfer learning’ approach, which is gaining popularity in artificial intelligence, they started by taking a model pre-trained on a public resource for computer vision that contains over a million common images, including cats, dogs, gadgets, etc. Then they fine-tuned the model to their specific task, which made it possible to differentiate between cell microscopy images.
“It teaches the network how to recognise images in a process similar to how a human learns during early childhood,” Theodore says. “The network then knows how to work with the images. And we kept fine-tuning the network so that it could find nuances in the images for our specific task.”
The key idea for fine-tuning the network was to partition all the cell images into four categories, loosely resembling cell cycle phases, and forcing the network to reproduce these differences. Surprisingly, it helped the network to reproduce the subtle differences with better accuracy. Finally, the network coded each cell with just four numbers that could identify the differences in the cells as a result of moving through the cell cycle process, representing the whole population of cells in just one picture.
“It was a beautiful reconstruction of the circular trajectory of the cell cycle, with the cells positioned along this trajectory,” Theodore says. “We validated it. We used the cell cycle-specific fluorescence markers to colour-code the cells according to their phases, and we saw that, yes, this organisation closely follows the cell cycle oscillation.”
The general method could be applied to other cells and could benefit scientists studying cancer development. “A hallmark of cancer cells is excessive proliferation. That means cells start dividing very quickly, so we need to understand not just why this happened but also how and when it happened,” Theodore says. “With the help of regular microscopy, without the need for any markers or genetic engineering, replacing them with the powerful capacities of a deep learning approach, this research can help pinpoint each cell’s own progression through the cell cycle or its ‘cell cycle time.’”


