Catching the cap-snatcher
Structural analysis opens the way to new anti-influenza drugs
Researchers at the European Molecular Biology Laboratory (EMBL) in Grenoble, France, have determined the detailed 3-dimensional structure of part of the flu virus’ RNA polymerase, an enzyme that is crucial for influenza virus replication. This important finding is published today in PLoS Pathogens. The research was done on the 2009 pandemic influenza strain but it will help scientists to design innovative drugs against all the different influenza strains, and potentially lead to a new class of anti-flu drugs in the next 5-10 years.
The scientists focused on the endonuclease part of the viral RNA polymerase. The endonuclease is responsible for a unique mechanism called ‘cap-snatching’ that allows the virus to trick its host cell into producing viral proteins. In human cells the translation of messenger RNA (mRNA) strands into proteins requires a special structure, called the “cap”, at the beginning of each mRNA. When the influenza virus infects a host cell its endonuclease “snatches” that cap from the cell’s own mRNA. Another part of its RNA polymerase then uses it as the starting point for synthesizing viral mRNA. With the correct cap structure at the beginning, viral mRNA can then hijack the protein-production machinery of the infected cell to make viral proteins, which assemble into new viruses that will spread the infection.
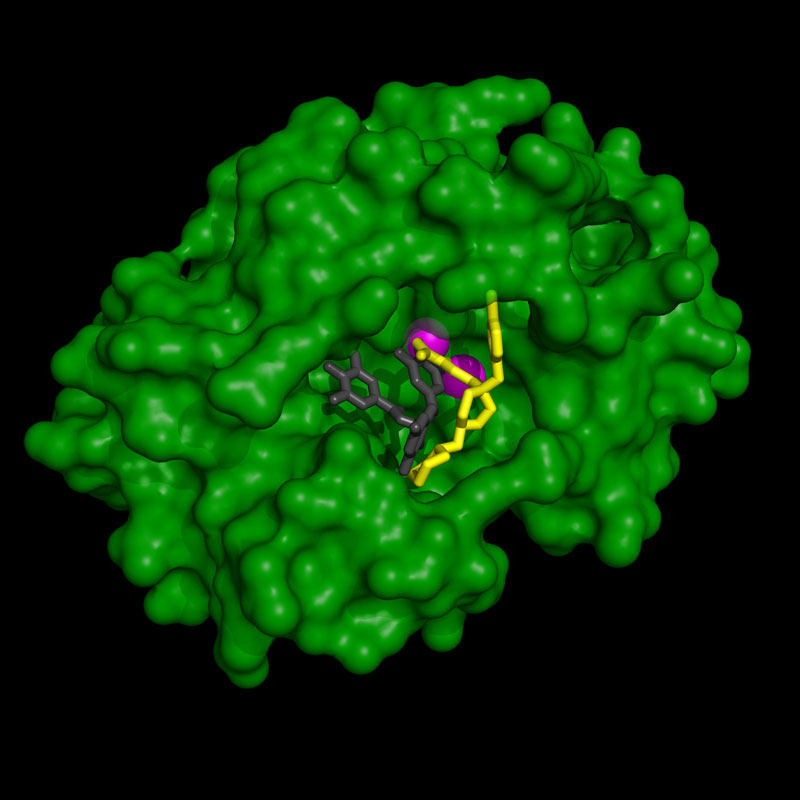
The team led by Stephen Cusack, Head of EMBL Grenoble, analyzed crystals of endonuclease from the 2009 pandemic influenza strain using the high intensity X-ray beams at the European Synchrotron Radiation Facility (ESRF). The researchers were able to determine the 3D atomic structure of the enzyme and to visualize how several different small molecule inhibitors bind to and block its active site. If the active site of the endonuclease is blocked by an inhibitor the enzyme cannot bind its normal substrate, the host cell mRNA, and viral replication is prevented.
The active site of the endonuclease is shaped like a cave with two metal ions at the bottom. Cusack and colleagues found that all the inhibitors they studied bind to those two metal ions but, depending on their shapes, different inhibitors bind differently to the amino-acids of the cave’s walls.
“Based on this detailed structural information we can now design new synthetic chemicals which bind even more tightly to the endonuclease active site and thus will potentially be more potent inhibitors of influenza virus replication,” explains Stephen Cusack. “We can even try to build in anti-drug resistance by making sure the inhibitors only contact those amino acids that the virus cannot mutate since they are essential for the normal activity of the polymerase.”
Because the cap-snatching mechanism is common to all influenza strains, new potent endonuclease inhibitors should be effective against seasonal flu, novel pandemic strains or highly pathogenic H5N1 bird flu. EMBL scientists are working with EMBL’s spin-off company Savira pharmaceuticals, in partnership with Roche, to further develop influenza inhibitors. Promising candidates will be tested first for efficacy in cell culture, ultimately moving into clinical trials on humans.
This research was partly funded by the European commission, through the FP7 research grant awarded to the FluPharm project.
Further information:
More on the collaboration with Savira pharmaceuticals
Article abstract
It is generally recognised that novel antiviral drugs, less prone to resistance, would be a desirable alternative to current drug options in order to be able to treat potentially serious influenza infections. The viral polymerase, which performs transcription and replication of the RNA genome, is an attractive target for antiviral drugs since potent polymerase inhibitors could directly stop viral replication at an early stage. Recent structural studies on functional domains of the heterotrimeric polymerase, which comprises subunits PA, PB1 and PB2, open the way to a structure based approach to optimise inhibitors of viral replication. In particular, the unique cap-snatching mechanism of viral transcription can be inhibited by targeting either the PB2 cap-binding or PA endonuclease domains. Here we describe high resolution X-ray cocrystal structures of the 2009 pandemic H1N1 (pH1N1) PA endonuclease domain with a series of specific inhibitors, including four diketo compounds and a green tea catechin, all of which chelate the two critical manganese ions in the active site of the enzyme. Comparison of the binding mode of the different compounds and that of a mononucleotide phosphate highlights, firstly, how different substituent groups on the basic metal binding scaffold can be orientated to bind in distinct sub-pockets within the active site cavity, and secondly, the plasticity of certain structural elements of the active site cavity, which result in induced fit binding. These results will be important in optimising the design of more potent inhibitors targeting the cap-snatching endonuclease activity of influenza virus polymerase.
Press Coverage
- THE MAIL ON SUNDAY (UK) , 2 AUGUST 2012New anti-flu drugs could be on the market within years after scientists create 3D map of part of the virus



