
Read the latest Issue
Microbes drive almost every food chain on Earth, using just about anything for energy and interacting with one another in countless ways. But how on Earth do they work?
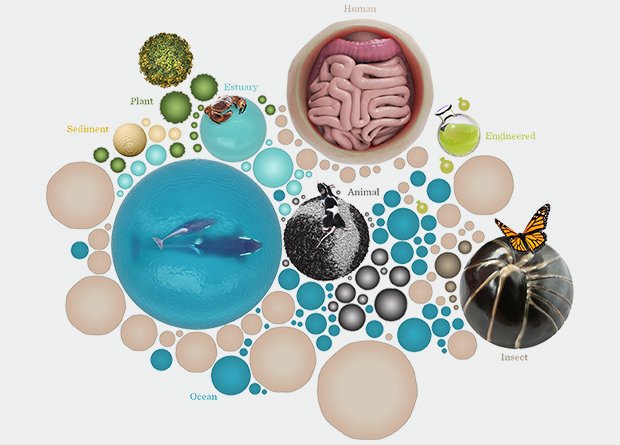
In the seas, in the soil, on our skin, in our gut – even in sewage treatment plants – microbes play pivotal roles in health and disease. To decipher how they work and how they influence each other, their hosts and the environment, scientists throughout the world are sharing and comparing vast amounts of information, all supported by EMBL-EBI.
A microbiome is an ecosystem of microbes: bacteria, viruses and other creatures invisible to the naked eye. Microbes are masters of their environment, growing everywhere from icy permafrost to boiling deep-sea vents, and influencing more of the living world than you might imagine.
Microbes are the great nitrogen fixers in our soil – in fact, with the right balance of them you can reduce the need for fertiliser to grow crops. They play a crucial role in our digestive systems – as anyone who has suffered an upset stomach after taking antibiotics knows from personal experience. They rule the seven seas, too – the microbiota in our oceans play a vital role in absorbing carbon dioxide, helping to sustain a healthy atmosphere.
Every one of us depends on these ecosystems to stay alive, so we have a lot to gain from understanding how they work, fall apart and recover. For example, if you spray detergent to clean up an oil spill, you might break up the oil nicely but the detergent will stick around in the environment, potentially affecting marine life forms – effectively, you would be solving one problem only to create another.
Alternatively, you could use the right combination of microbes to remove that oil by breaking it down into harmless compounds like sugar. That would make it much easier to restore the microenvironment to its original state.
But how do you do that?
The tricky thing is that most microbes are very difficult to study outside the context of their communities. This means scientists can’t grow the microbes in a Petri dish and analyse them in isolation. Instead, they must study the whole environment, looking at all the genetic material in a sample and trying to tease out what organisms it came from.
Describing microbiomes in detail – making a community directory – is really difficult, as the individuals aren’t easy to pinpoint.
So how do you find out who’s who, what they’re eating, and how they function? How can you pin down a beneficial chain of events in a given microbiome, such that you can manipulate it – returning it to health after the chain has been interrupted – or turn it to some new use, like sewage treatment or biofuel production?
If we know what makes a microbiome function, we can figure out how to keep it healthy, or fix it when it’s disrupted.
Humans are pretty good at sequencing DNA these days – everything from whole genomes of people and plants to pathogens lurking in a hospital environment, identified on-the-fly.
But DNA sequences are pretty meaningless unless you can compare them to a reference. That can tell you whether it’s a known sequence (which is pretty important) and, if so, what it is and what’s special about it. Then again, you might discover that a sequence is novel, because there is no reference in the public archive.
So far, so good. But what if you want to sequence a spoonful of soil, a bucket of seawater, a stool sample, a scraping of skin, a little patch of the forest floor – each of which has millions of microbes and an untold number of DNA sequences? You’ll end up with a huge amount of new information, all of which you want to compare to a reference archive at once.
Does that even work?
Yes, it does work. Studying the DNA sequences of a whole environmental sample, whether it’s from the sea or your skin, is called ‘metagenomics’ – literally, the study of genomes within genomes.
“You can take a mixture of microbes from an environment and use computational approaches to identify what’s in there, skipping the part where you separate out and culture individual viruses and bacteria,” explains Guy Cochrane, who runs the European Nucleotide Archive – a major source of reference sequences and, as part of the International Nucleotide Sequencing Consortium (INSDC), the engine driving much of genomics research today. “Collecting, sequencing and comparing samples is something scientists are pretty good at, so we can provide a strong start towards understanding what these communities are doing biochemically,” he adds.
“With environmental samples, you have to sequence so much stuff!” says Rob Finn, who runs EMBL-EBI’s Metagenomics data service, which contains tens of thousands of analysed datasets representing genetic material from environmental samples.
“Comparing the sequences found in one microbiome might be the equivalent to analysing a few thousand human genomes – that is simply a staggering amount of information. Our Metagenomics service helps people discover what is actually in their samples, and classify things that are being described for the first time.”
“You can compare the DNA in a sample of contaminated soil to a clean sample from roughly the same place, or healthy gut to a diseased one, look for the differences and make some reasonable assumptions about what those genes are doing,” adds Finn. “We need to look beyond that, so we can start to see what makes one microbiome thrive and another languish. If we can manipulate these environments, we can start to solve some serious problems.”
Anticipating disease outbreaks is an incredibly important application for metagenomics. The COMPARE project, an international pathogen-surveillance network, uses genome sequencing technology and rapid data sharing to speed up the detection of infectious disease outbreaks.
COMPARE is working to set up monitoring of sewage treatment plants in cities throughout the world, looking at which bacteria are circulating in a population, and what infectious diseases may be brewing.
“In COMPARE, our colleagues are using advanced methods, including some developed in Rob’s team, sharing data as it is generated and using our cloud compute for some of the analyses. So they don’t have to worry about the infrastructure – they can get on with figuring out how these things are moving from place to place and evolving,” says Cochrane.
Microbiome research, like cancer research, is very, very broad and endlessly complex. It’s a poster child for ‘interdisciplinary research’, demanding expertise in many areas of science.
“Studying the microbiome covers so many different areas that it brings together a hugely diverse community,” says Finn. “The people involved in international microbiome projects bring a lot of different knowledge to the table: microbiology, ecology, genetics, metabolomics, clinical research, healthcare, ecology, biodiversity… They’re specialists, bioinformaticians, citizen scientists, participants at all levels, and they need the right tools to get on with their part of the work.”
It is endlessly fascinating – and useful – to see how life processes are connected and influence one another.
The human microbiome can be considered akin to the brain or another organ. After all, our bodies contain a substantial number of both parasites (the agents of disease) and symbionts (providing essential functions). At EMBL, several research groups are collaborating to understand the human microbiome and its role in our lives.
“Microbiomes are the focus of intense research at EMBL,” says Peer Bork, whose research group explores how the environment in the human gut influences how a person responds to a drug. “My group has been exploring microbial functions in the gut since 2004, and thanks to the Tara Oceans expeditions, we’ve also had the opportunity to compare these communities inside humans with those on the surface of the sea. It is endlessly fascinating – and useful – to see how life processes are connected and influence one another.”
The importance of plankton in maintaining the Earth’s climate cannot be understated – their communities absorb a staggering volume of CO2 from the atmosphere and release oxygen in exchange. Yet only a small fraction of these life forms have been classified and analysed.
To address this gap, EMBL scientists initiated and led the Tara Oceans expeditions: an unprecedented effort that resulted in 35,000 samples of seawater, each of which contained millions of small organisms. The samples, studied using advanced microscopy from EMBL, were sequenced at Genoscope in France, generating over 7000 datasets. Teams of researchers all over the world delved into this richly described collection, bringing to light 40 million novel genes and a raft of discoveries about life in the world’s oceans.
Where can you find Tara Oceans data, and other marine datasets? At EMBL-EBI, of course.
“We’ve worked with sequencing experts at Genoscope in France, our research colleagues at EMBL Heidelberg, data providers at Pangaea in Germany and many, many others to make Tara Oceans data available to everyone,” says Cochrane. “This was an extensive sampling programme that was well organised from the start, so we have rich information about their chemistry and environmental context. That makes these datasets incredibly valuable over the very long term, because people can use them over and over again, in new ways, well into the future.”
Comparing Tara Oceans, Ocean Sampling Day and Malaspina marine datasets has already provided insights into the profound differences between the microbiomes of deep, shallow and coastal marine waters.
Genome technology is advancing so quickly that it can be difficult to keep pace. EMBL-EBI teams work with researchers, data providers, technology companies and even biotech start-ups to make sure the right machinery is in place to keep science moving.
“Our goal is building knowledge, not just gathering data,” says Paul Kersey, who leads the Ensembl Genomes project at EMBL-EBI. “If you want to understand something as complex as a microbiome, you’ve got to have the best possible data on every level, and provide intelligent ways to cross-link and query them. It only works if you can present new information in the context of what is already known.
“This is precisely what the Ensembl Genomes service does. We bring together all the knowledge gained about the genomes of microbial life forms – linking historically well-studied species with those only recently discovered.”
Our job is to normalise the data […] and connect it all up so that people can slice across it in interesting ways.
“It’s all very exciting, and scientists are easily tempted to jump in and gather data very quickly, using the latest kit, and upload it into an archive as an afterthought,” says Cochrane.
“Our job is to normalise the data – no matter what technology was used – and connect it all up so that people can slice across it in interesting ways. That’s how you get researchers finding novel genes that influence diseases like Crohn’s, or discovering cold-loving enzymes that can take the place of detergents,” adds Finn.
Microbiome research is very new, and changing rapidly as new technologies and approaches widen the range of questions we can address. At EMBL-EBI, the Finn team is collaborating with a company called BioCatalysts, using metagenomics to discover novel enzymes that could have industrial applications. Discovering these enzymes using a data-driven approach rather than carrying out chemical synthesis experiments would curb the generation of chemical waste.
Want to know more about life in the seas? On 21 June, every year, the Micro B3 project invites sailors, skippers and anyone working in marine research to participate in Ocean Sampling Day (OSD), a simultaneous sampling campaign of the world’s oceans, from the subtropical waters around Hawaii to the rather brisk rapids of the Fram Strait in the Arctic. By taking samples at the same time every year, the hundreds of scientists taking part in OSD are describing microbial diversity and function, which at present is very poorly understood, and identifying trends over time. Not working on a boat or in marine science? The OSD Citizen Science campaign welcomes everyone to collect marine microbes and environmental data to help scientists piece together a better understanding of the world’s oceans.
Want to know more about the human gut microbiome? The my.microbes project (initiated by EMBL scientists), the British Gut Project and the American Gut project all welcome citizen scientists to provide samples and information about diet and lifestyle. These projects, taken together, will help map the gut microbes of human populations throughout the world, building an understanding of how lifestyle choices and diet influence our microbiomes, and therefore our health.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive