
Read the latest Issue
EMBL Grenoble scientists present important insights into how DNA is organised during the cell cycle.
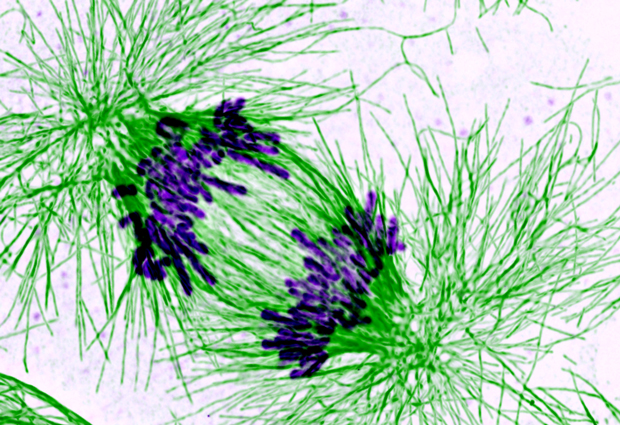
From yeast to human, from birth to death – our cells are continually going through a cycle of growth and division. Central to this cycle is DNA – each time it must be correctly duplicated and distributed into the daughter cells when the cells divide. Failure to do so can result in cell death, cancer or infertility. A molecular machine crucial for the accurate implementation of this process is cohesin – a ring of proteins that latches onto the cell’s DNA ensuring that identical copies of the DNA stay together and divide correctly into the daughter cells. But just how cohesin keeps everything together is not completely understood. “What we would really like to understand is how closure of this ring is regulated and how it stays stable for long periods of time” explains EMBL Grenoble group leader Daniel Panne. Kyle Muir, a PhD student in Panne’s group, has now succeeded in shedding some light on the process.
In a paper published recently in Cell Reports, the researchers present the 3D atomic structure of one of the cohesin ring proteins (Scc1) bound together with a protein called Pds5. It has long been acknowledged that Pds5 is important for stabilizing the cohesin ring and, paradoxically, promoting its break up, but just how is not known. “Our structure shows that there is a really small, but essential, interface for Pds5 to bind to the cohesin ring” explains Muir, “and this appears to be conserved across the animal kingdom – so all organisms from yeast to man have this exact same interface between the two proteins”. This highlights the importance of Pds5 for the functioning of cohesin. Indeed additional cell biology experiments carried out in the lab of Christian Haering at EMBL Heidelberg to disrupt this interface resulted in the cells dying because they were unable to maintain chromosome pairs. Although details of the whole process remain elusive, the researchers believe Pds5 may be central for the recruitment of additional proteins which carry out different tasks during the cohesin cycle of establishment, stability and break-up. “The structure also reveals aspects of Pds5 that might enable it to control closure of the cohesin ring, which will allow researchers to design specific experiments to dissect the various parts of this crucial process” concludes Muir.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive