
Lars M. Steinmetz
Associate Group Leader / Director of Life Science Alliance
EditSystems genetics and precision health

Abstract:
In CARDIOREPAIR, we aim to identify and repair disease-causing variants in dilated cardiomyopathy (DCM), the major contributor to heart failure. We focus on the gene RBM20 mutated in patients with a highly penetrant and aggressive form of familial DCM. We will create a comprehensive map of all possible RBM20 variants that can cause DCM and develop a therapeutic approach for mutations with the strongest effect size. Enabled by a multitude of new technologies developed in our groups, the primary goal is to characterize the complete spectrum of amino acid mutations in RBM20 by high-throughput saturation mutagenesis screens. Combined with functional readouts and multi-omics analysis of downstream processes, we will classify and score the pathogenicity of each individual mutant (objective 1). To this end, we will cover both the known DCM-causing RBM20 variants, as well as those that have not been identified in patients yet. Each mutant, represented by a typical transcriptomic, proteomic, phosphoproteomic, and microscopic fingerprint, will guide us in finding novel class-specific therapeutic strategies to revert the deviant phenotype back to the healthy state. For a subset of mutants representing each identified mutation class, we will generate mouse models and investigate changes in heart physiology and ultrastructure linked to the altered ‘ome’ profile. Our second goal is to establish a class-specific therapeutic approach for the treatment of patients harboring RBM20 mutations (objective 2). We will implement our advances in muscle-specific gene editing, focusing on prime editing and nanobody-guided approaches to specifically tackle the mutations leading to RBM20 translocation. This proposal serves as a blueprint for going from variant identification to therapy in an accelerated fashion by harnessing and combining the power of high- throughput functional genomics and bioengineering and therefore is widely applicable to other cardiovascular diseases (CVD).


Associate Group Leader / Director of Life Science Alliance
EditLars studied molecular biophysics and biochemistry at Yale University and conducted his Ph.D. research on genome-wide approaches to study gene function and natural phenotypic diversity at Stanford University with Ron Davis. Lars started his own group 2003 at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, focused on applying functional genomic approaches and high-throughput methods to study complex traits, transcription and the mitochondrial organelle at a systems level. In parallel, he maintained a focused group at the Stanford Genome Technology Center working on technology development.
Kai studied molecular and cellular biology and did his PhD at the ZMBH in Heidelberg. During his PhD, he developed a method to detect interactions between nascent proteins translated by adjacent ribosomes, and reveled the prevalence of this co-translational protein assembly mechanism in human cells. In his postdoc he is now focusing on high-throughput genotype-phenotype couplings. He is the project organizer of the CardioRepair project.

Research Scientist
EditJulia studied molecular biology, and obtained her joined PhD degree from EMBL and Heidelberg University in 2023. During her PhD, she applied functional genomics together with image enabled cell sorting to identify the mechanism of RBM20 mislocalization in dilated cardiomyopathy (DCM) which yielded a novel therapeutic avenue. Now she continues to lead projects aimed at identifying novel approaches to treat RBM20-mediated DCM.

Postdoctoral Fellow
EditCarlos completed his PhD at the Max Planck Institute of Immunobiology and Epigenetics in Freiburg, where he developed experimental and computational transcriptomic approaches to study RNA-binding proteins. Currently, he is a postdoctoral fellow in the Steinmetz lab at EMBL, contributing to the EIC project.

Predoctoral Fellow
EditLinda studied Genetics and Genomics at Helsinki University and is currently employed as a Predoctoral Fellow at EMBL Heidelberg. Her project focuses on expanding functional readout strategies for high-throughput variant effect mapping, aiming to advance personalised health approaches.

Predoctoral Fellow
EditAnastasiia studied Molecular and Cellular Biology at the University of Glasgow and completed her masters project at the Francis Crick Institute. She is currently working as a predoctoral fellow at the EMBL Heidelberg focusing her research on the in-depth dissection of CRISPR editing in vivo using spatial transcriptomics and single cell sequencing methods.

Lab Manager
EditSandra

Group leader
Gil is the Director of the Institute of Synthetic Biomedicine at Helmholtz München and the Chair of Neurobiological Engineering at Technical University of Munich (TUM), where several research teams are integrating protein design, genome editing, and mammalian models to develop next-generation biotechnologies for molecular sensing and control of cellular processes, with the ultimate goal of advancing future cell therapies.

Group leader
Jeff studied Molecular Biotechnology and did his Ph.D. at TUM/Helmholtz Munich; during his PhD, he developed new genome engineering/gene therapy tools as well as non-invasive gene reporter systems for protein-isoforms and non-coding RNA. He is now the scientific lead for the Munich side of the EIC project.

Predoctoral Fellow
Julian studied Molecular Biotechnology at TUM and is currently finishing his PhD co-developing tools for cell and gene therapy and tools for post-translational correction of cellular abnormalities.

Predoctoral Fellow
Niklas studied Molecular Biotechnology at TUM and is currently completing his PhD at TUM/Helmholtz Munich co-developing non-invasive transcriptome reporter systems and related cell and gene therapy technology.

Technician
Tobi studied Molecular Biotechnology at the TUM and is helping the EIC Munich team in his role as a Lab Manager and with experiments in mammalian cell systems.

Julia presented her most recent findings about the interaction between RBM20 and TNPO3

Unraveling the Mechanism RBM20 Mislocalization in Dilated Cardiomyopathy Reveals a Novel Therapeutic Avenue
Numerous human diseases are linked to incorrect protein localization within cells. The development of effective therapies targeting such conditions depends on a deep understanding of their specific molecular mechanisms. In this study, we focus on deciphering the mechanism of pathogenic RBM20 variants, which lead to aggressive types of dilated cardiomyopathy (DCM). Alterations in the RS-domain of RBM20 disrupt mRNA alternative splicing regulation, and lead to cytoplasmic RBM20 mislocalization in a form of RNP granules. To discover factors that regulate RBM20’s transport to the nucleus in an unbiased way, we conducted a genome-wide CRISPR-Cas9 screening and employed image-enabled cell sorting (ICS) to isolate cells exhibiting RBM20 mislocalization. We identified TNPO3 as the primary transporter of RBM20 into the nucleus and demonstrated a direct interaction between the two proteins. We confirmed that mutations in the RS-domain disrupt the TNPO3-RBM20 connection, leading to RBM20 mislocalization. Further, we investigated the splice regulatory function and protein interactions of RBM20 variants upon their natural or experimentally induced relocation to the nucleus. Our findings reveal that relocating RBM20 to the nucleus restored its alternative splicing function and resolves the accumulation of cytoplasmic RNP granules. This suggests that RS-domain mutations in RBM20 maintain a level of their regulatory capacity on splicing, with the disease’s progression primarily driven by the failure of these variants to efficiently localise the nucleus. We are further developing potential therapeutic approaches such as artificial binders to relocalise RBM20 variants. In summary, our work on the mislocalization of RBM20 opens up a novel therapeutic avenue for patients with RBM20-related DCM.

Kai presented the most recent results about our high-throughput variant effect mapping via single cell phenotyping.

Deciphering the mechanism of RBM20 mislocalization in dilated cardiomyopathy identifies a novel therapeutic avenue.
Many human pathologies are associated with aberrant protein localization. Successful development of targeted therapies for such diseases requires understanding of their precise molecular pathomechanisms. Here, we address the mechanism of pathogenic RBM20 variants that cause severe forms of dilated cardiomyopathy. Mutations in the RS-domain of RBM20lead to impairment of mRNA splicing regulation, cytoplasmic RBM20 mislocalization, and formation of RNP granules.To identify regulators of RBM20 nuclear import in an unbiased way, we performed a genome-wide CRISPR-Cas9 screen, and used image-enabled cell sorting (ICS) to isolate cells with mislocalized RBM20. We identified TNPO3 as the main nuclear importer of RBM20, and showed the direct binding between both proteins. We validated that pathogenic RS-domain mutations abolish the TNPO3-RBM20 interaction, which triggers mislocalization of RBM20. We analyzed splice regulatory activity and protein interactors of RBM20 variants upon their physiological or induced nuclear re-localization.We showed that nuclear relocalization resulted in restoration of alternative splicing and clearance of cytoplasmic RNP granules. This indicates that RS-domain RBM20 variants at least partially retain their splicing regulatory activity, and that the main driver of the disease progression is their inability to be efficiently imported into the nucleus.


The Steinmetz lab and the Westmeyer lab presented currently ongoing RBM20-DCM related projects during the annual DCM meeting (organized by Lars Steinmetz) and used the time to exchange ideas and discuss the progress with all participating labs.
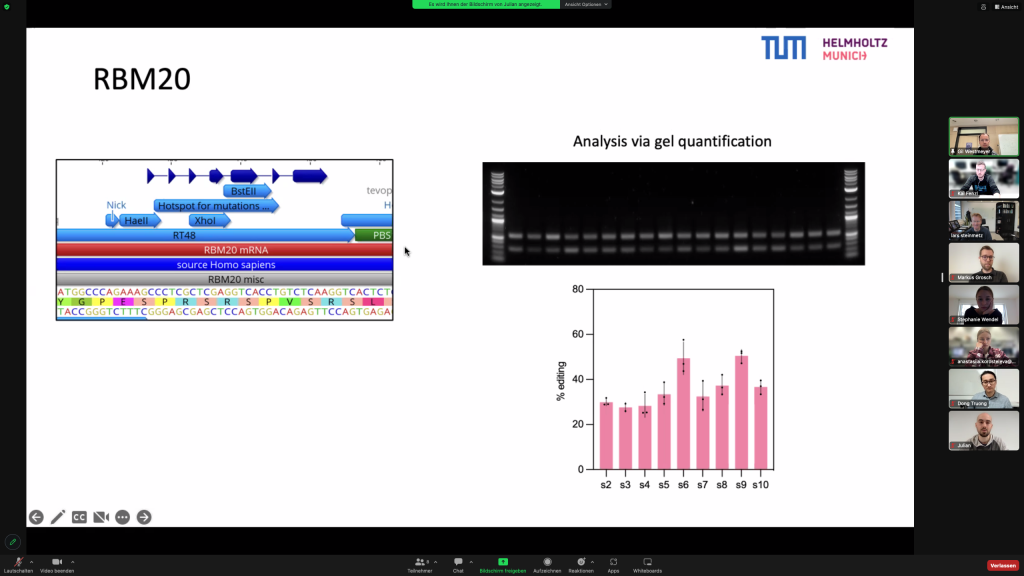
The Steinmetz lab and the Westmeyer lab presented currently ongoing RBM20-DCM related projects during that meeting, exchanged ideas and discussed the next planned collaborative steps for the CardioRepair grant.
HMGU: https://www.helmholtz-munich.de/en/newsroom/news-all/artikel/expanding-the-gene-editing-toolbox
TUM: https://www.bioengineering.tum.de/en/news/details/expanding-the-gene-editing-toolbox
Truong, DJ.J., Geilenkeuser, J., Wendel, S.V. et al. Exonuclease-enhanced prime editors. Nat Methods21, 455–464 (2024). https://doi.org/10.1038/s41592-023-02162-w

