
Eva Kowalinski
Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-9785-9027
EditStructural biology of macromolecular protein-RNA complexes

Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-9785-9027
EditThe group has a keen interest in gene expression regulation through RNA processing pathways. Post-transcriptional chemical RNA modifications are ubiquitous (tRNA, mRNA, rRNA, snoRNA, etc.) and occur in all kingdoms of life. For example, eukaryotic tRNAs contain on average 13 modifications per molecule that assure translation fidelity and efficiency. Modifications to mRNAs have effects on folding, solubility, transcript stability, splicing, and RNA localisation. Thus, the so-called epitranscriptome adds an additional layer of information to the message. This layer is reversible in response to cellular stress, metabolic changes, or developmental stage. In the Kowalinski group, we are investigating macromolecular complexes that modify RNA or recognise modified RNAs. We solve structures of these complexes by single-particle cryo-electron microscopy (cryo-EM) and X-ray crystallography to understand the following questions: how exactly is a specific RNA selected for modification? How is this process regulated? How does a modified RNA differ in its binding to downstream effectors?
In the future, the Kowalinski group wants to gain insight into the mechanisms that specify certain RNAs for modification and how the editing is controlled. To this end, we will not only reconstitute recombinant complexes in vitro but also purify native endogenous complexes for structural investigation. We use X-ray crystallography, cryo-EM, and scattering techniques like small-angle X-ray scattering (SAXS), combined with biophysical methods, biochemical assays, computational structure prediction and cell biology to assess the structure–function relationship within these complexes. RNA modifications are relevant to diseases like cancer but also play a role in infectious diseases such as trypanosomiasis and leishmaniasis. In the future, we hope to be able to use our knowledge for structure-aided drug design.

Rutuja Yelmar, predoctoral Fellow at EMBL Grenoble in the Kowalinski group, talks about her passion for research, daily inspiration, and the importance of collaboration in achieving her goals.
Edit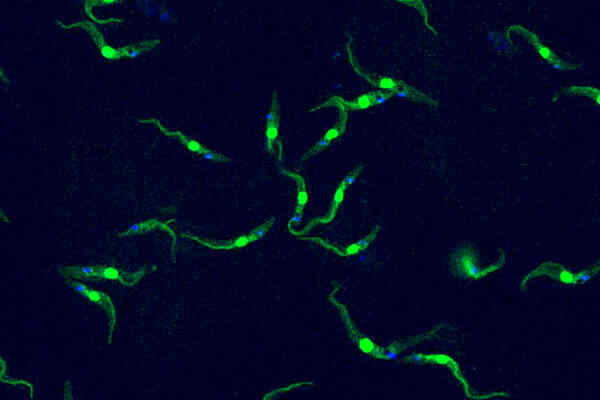
This single-celled organism the size of a dust particle is capable of causing deadly tropical diseases in both humans and livestock –Trypanosoma brucei, in an image by Luciano Dolce from EMBL.
Edit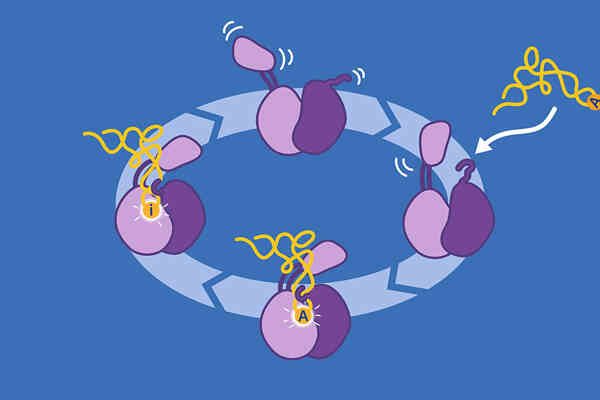
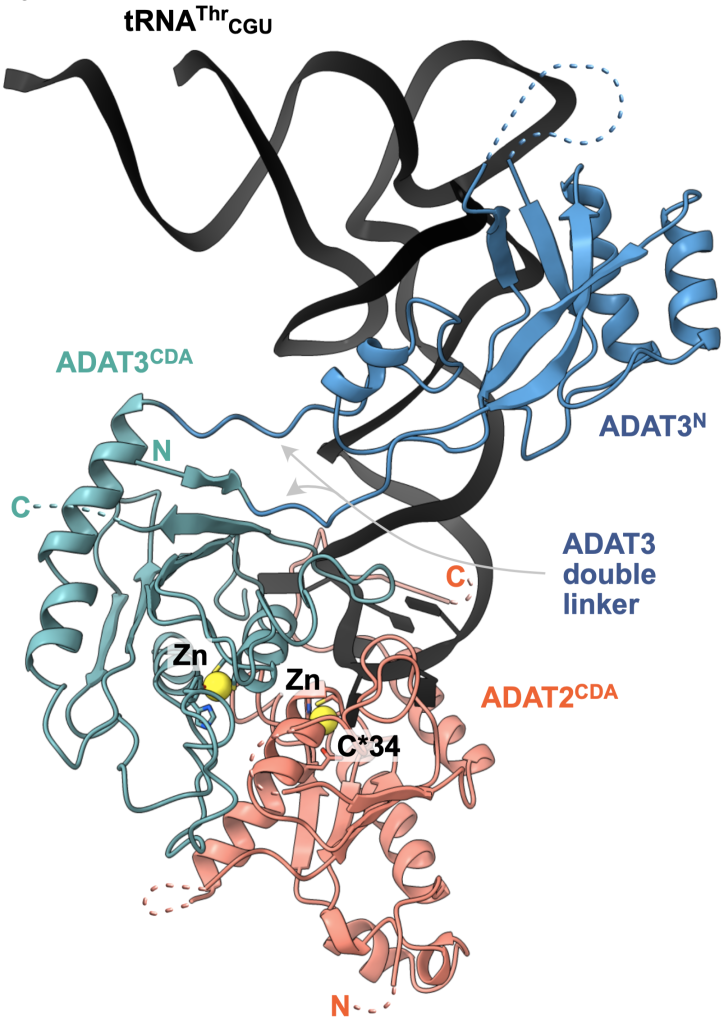
EMBL Grenoble scientists provide new insights into the function of an essential RNA editing enzyme.
Edit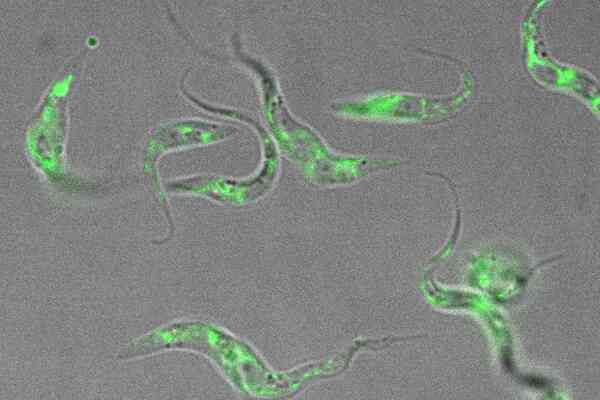
Members of the EMBL community are working to improve our understanding of the parasites that cause malaria and sleeping sickness
Edit
New group leader based in Grenoble aims to unveil the mechanisms of RNA editing
Edit