
Eva Kowalinski
Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-9785-9027
EditStructural biology of macromolecular protein-RNA complexes

Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-9785-9027
EditThe Kowalinski group aims to understand gene expression regulation through RNA processing pathways. We investigate novel concepts in eukaryotic RNA processing, including mechanisms of RNA editing and chemical modification. We focus on both fundamental and applied questions. How is a specific RNA selected for processing? How is RNA processing differentially regulated in health and disease? Can we inhibit RNA modification enzymes with small molecules, for example, to treat cancer? Can we use RNA processing machines as tools for transcriptome editing?
We use a multidisciplinary approach integrating diverse structural biology methods (single particle cryo-electron microscopy, in situ cryo-electron tomography, and X-ray crystallography) with biochemistry, biophysical methods, computational structure prediction, and cell biology. Our goal is to understand the mechanisms of RNA-processing molecular machines from atomic to cellular resolution. We employ recombinant approaches and prepare samples directly from cells or reaction mixes, with the ultimate goal of elucidating the mechanics and dynamics of our target complexes within cells.
Trypanosomes are unicellular protists that diverged from a common ancestor over 500 million years ago. They are excellent models for studying evolutionary diversity. Cutting-edge genetic tools are available in lab strains of Trypanosoma brucei. Beyond fundamental research, Trypanosoma brucei can be used as a model organism to discover novel anti-infective drugs against related parasites that affect humans and animals.
Trypanosomes undergo several developmental stages during their life cycle as they adapt to the environment in their host. Their gene expression is controlled primarily by regulating the processing, stability, nuclear export, translation, and degradation of RNAs.
Our research questions: What makes RNA processing and trans-splicing so efficient in trypanosomes? How do trans-spliceosomes differ from conventional (cis-) spliceosomes? With this knowledge, can we induce efficient trans-splicing in cis-spliceosomes? How is alternative trans-splicing regulated? What is the role of the sub-nuclear compartments in organising RNA processing? What molecular mechanisms govern mitochondrial mRNA editing?
Insight: We have revealed the architecture of the unusual tetrameric nuclear cap-binding complex of Trypanosoma brucei, which is essential for trans-splicing. We demonstrated that this cap-binding complex requires the canonical m7G methylated RNA cap structure (cap-0) for binding, but does not require the subsequent methylations of the atypical cap-4 structure present in trypanosomes (Bernhard et al., 2025).
Insight: Large multi-subunit complexes coordinate mitochondrial template-based mRNA editing in trypanosomes to transform cryptic mRNAs for respiratory chain complexes into protein-coding transcripts. By using cryo-EM and biochemistry, we established that RESC1–RESC2, a central module of the RNA editing substrate-binding complex (RESC), is the protective 5′-end binding complex of gRNAs and that it specifically selects gRNAs from the mitochondrial RNA pool (Dolce et al. 2023).
Chemical modifications of mRNA, rRNA, and tRNA – the epitranscriptome – shape structure, function, and localization of RNAs and add to the complexity of gene regulation in eukaryotes. The intricate interplay between modifications of the tRNA anticodon loop and the mRNA codon ensures the efficiency and fidelity of protein translation by the ribosome. The disruption of the healthy epitranscriptome causes cancers and neurological disorders.
Our research questions: How do RNA modification enzymes select their specific substrates? How are RNA modifications regulated in development, stress, or infection?
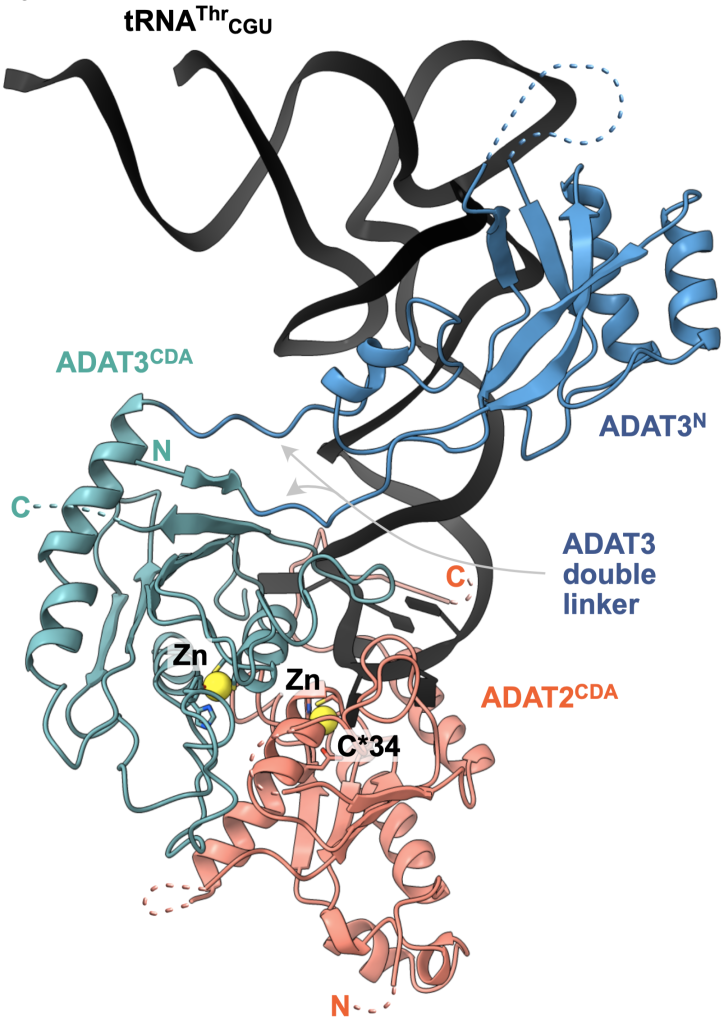
Insight: The conversion of adenosine to inosine in the wobble position of the tRNA anticodon by the ADAT deaminase is vital for eukaryotes and fine-tunes codon usage in certain cell types and developmental processes. Our cryo-EM structure and biochemical analyses revealed how the essential eukaryotic ‘wobble’ base deaminase ADAT2–ADAT3 utilizes flexible and intrinsically disordered protein motifs to recognize the geometric features of diverse tRNAs without deaminating off-target stem-loops (Dolce et al., 2022).
Insight: The N3-methylcytosine (m3C) modification of C32 in the tRNA anticodon loop in a subset of human tRNAs ensures error-free translation under stress conditions. We are studying the mechanisms of substrate recognition by the m3C ‘writer’ enzymes METTL6, METTL2, and METTL8 and evaluating their therapeutic potential. We demonstrated that METTL6 relies on seryl-tRNA synthetase (SerRS) to recognize and methylate its substrate tRNASer, and revealed the catalytic mechanism of m3C RNA methyltransferases (Throll et al., 2024; Aupič et al., 2025). Crucially, our cryo-EM complex structure included an endogenous tRNASer, allowing us to visualize chemical RNA modifications at atomic resolution and proving that this is possible using cryo-EM.

Eva Kowalinski received an ERC Consolidator Grant for her promising work on fundamental cellular mechanisms
2025

The Kowalinski group at EMBL Grenoble identified significant differences between the trypanosomal and human nuclear cap-binding complex, a key player in cellular RNA metabolism and a potential target …
Edit
EMBL Grenoble’s Kowalinski Group analysed the structure of an enzyme responsible for modifying tRNA molecules to fine-tune protein production. They discovered that to distinguish almost identical, yet…
Edit
Rutuja Yelmar, predoctoral Fellow at EMBL Grenoble in the Kowalinski group, talks about her passion for research, daily inspiration, and the importance of collaboration in achieving her goals.
Edit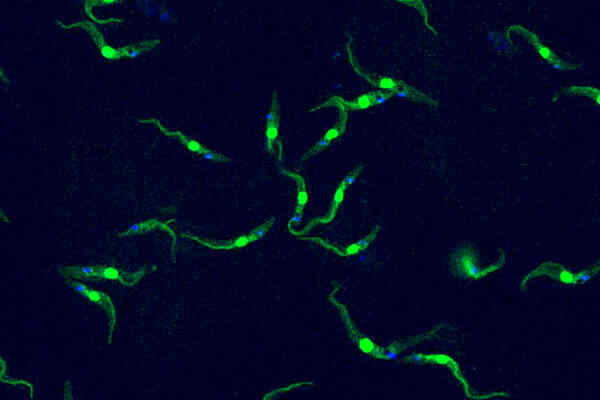
This single-celled organism the size of a dust particle is capable of causing deadly tropical diseases in both humans and livestock –Trypanosoma brucei, in an image by Luciano Dolce from EMBL.
Edit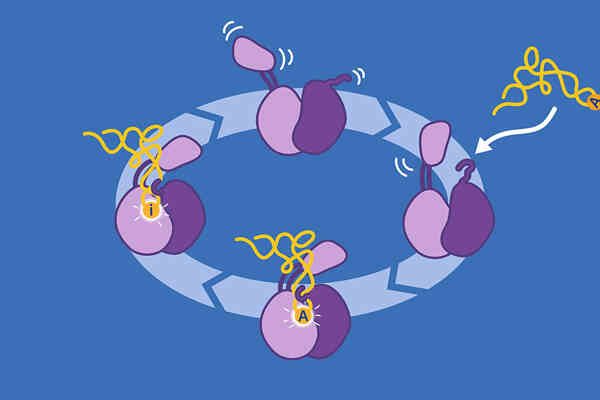
EMBL Grenoble scientists provide new insights into the function of an essential RNA editing enzyme.
Edit