Jan Ellenberg
Group Leader
Cell Biology and Biophysics Unit
EditCell division and nuclear organisation

Group Leader
Cell Biology and Biophysics Unit
EditOur overall goal is to elucidate the molecular and physical principles underlying cell division and nuclear organisation. We are developing a broad range of advanced fluorescence-based imaging technologies to assay the functions of the involved molecular machinery non-invasively and quantitatively, to automate imaging to address all its molecular components, and to computationally process image data to extract biochemical and biophysical parameters.
We have previously applied systems biology approaches to identify new cell division genes by RNAi-based screening of the entire human genome and we are now studying – in live cells and with high throughput – crucial protein functions, interactions and networks during cell division. We are combining automated single molecule-calibrated imaging and computational data analysis with advanced machine learning and modelling approaches to extend the first integrated protein atlas of the human dividing cell (video 1) (Cai et al., Nature 2018).
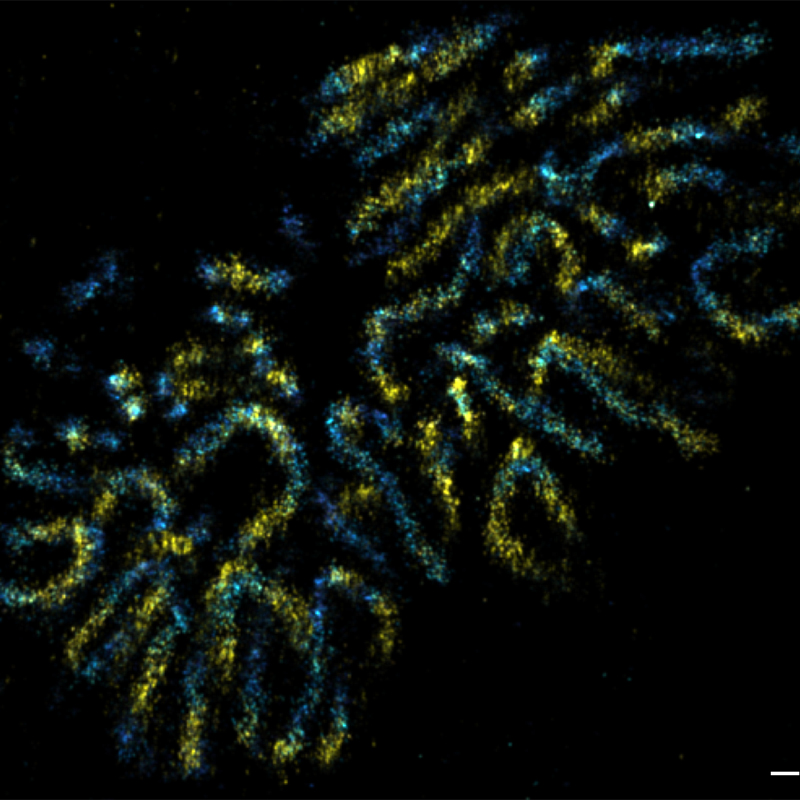
We also determined the positions of various nuclear pore complex (NPC) components and directly resolved the ring-like structure of the NPC by super-resolution microscopy (Szymborska et al., Science 2013). Applying correlative light and electron microscopy, we recently discovered fundamental differences in postmitotic and interphase NPC assembly (Otsuka et al., eLife 2016; Otsuka et al., Nat Mol Struct Biol 2018). Currently, we are dissecting those assembly mechanisms of the NPC molecularly, as well as studying the structure of the NPC’s dynamic components. We are also studying chromatin dynamics throughout the cell cycle, and unraveling the 3D architecture of the human genome in situ (Walther et al., J Cell Biol 2018; Xiang et al., J Cell Biol 2018).
By complete kinetochore tracking, we demonstrated that meiotic chromosome biorientation is highly error-prone (Kitajima et al., Cell 2011). We have now developed a gentle light-sheet-based microscope for high-throughput imaging of mouse embryos to enable systematic molecular analysis of early embryonic mitosis (Strnad et al., Nat Methods 2016). This allowed us to demonstrate that parental genomes are kept apart by a dual spindle in mouse zygotes (Reichmann et al., Science 2018).
We want to gain further comprehensive mechanistic insight into the division of human mitotic cells, to provide a biophysical basis for understanding nuclear organisation, and to establish methods for systematic analysis of the surprisingly error-prone first mitotic divisions of mammalian embryos.
To come to a structural understanding of nuclear organisation, we will explore and further improve correlative imaging approaches, combining live-cell confocal microscopy, super-resolution and electron tomography to unravel the structure and mechanisms of NPC assembly and disassembly.
In order to understand the function of the human genome, knowing the genome sequence alone is not sufficient. We are studying chromatin organisation and compaction, as well as the 4D human genome architecture by correlative imaging approaches, including live-cell imaging and several super-resolution techniques (Figure 1).
We will push light-sheet-based imaging technology development further to improve its light efficiency and resolution in order to establish a physiological molecular model for early mammalian development and infertility.