Multimodal Imaging combines the strength of several imaging techniques. For instance, Correlative Light and Electron Microscopy (CLEM) enables the targeting of a specific structure or event of interest and inspection at the nanometer scale, thereby bridging scales and resolutions.
In the EMCF, we offer several flavours of multimodal imaging, to achieve precise localization (using light microscopy or X-ray imaging) and to further study the ultrastructural context (using electron microscopy) .
On-section CLEM
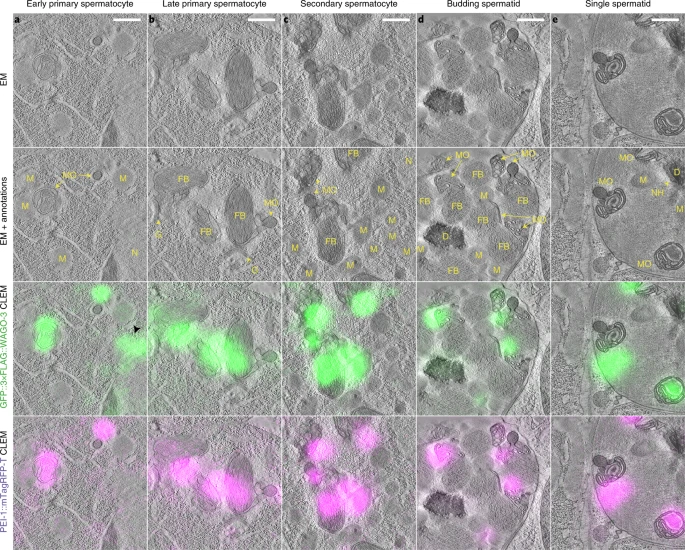
In-situ CLEM workflows retain the fluorescent signal of a labeled target protein (Nixon et al., Kukulski et al.). One can preserve the fluorescence by high-pressure freezing the specimen, capturing it in its native state, followed by freeze substitution using a low amount of heavy metals. Thick sections are cut (250 – 300nm thick) and placed on an EM grid allowing the very same regions on the section to be imaged by wide-field fluorescence microscopy and further by electron tomography. A detailed step-by-step tutorial on the method can be found here. Note that the same sample preparation protocol is the basis on volume CLEM, as detailed below.
Examples of recently studied organisms at the EMCF are C. elegans, D. melangoaster, S. Cerevisiae, and different mammalian cell lines.
References:
Nixon SJ, Webb RI, Floetenmeyer M, Schieber N, Lo HP, Parton RG., A single method for cryofixation and correlative light, electron microscopy and tomography of zebrafish embryos. Traffic. 2009 Feb;10(2):131-6.
Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA., Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011 Jan 10;192(1):111-9.
Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA., Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol. 2012;111:235-57.
Schreier, J., Dietz, S., Boermel, M. et al. Membrane-associated cytoplasmic granules carrying the Argonaute protein WAGO-3 enable paternal epigenetic inheritance in Caenorhabditis elegans. Nat Cell Biol 2022 Feb;24(2):217-229.
Live Cell CLEM
Live Cell CLEM enables the researcher to view the fluorescence signal in their cells in culture live with (fluorescence) light microscopy prior to EM sample preparation. Thus, a certain event or phenotype is identified and the sample quickly fixed thereafter by chemical fixation or high pressure freezing.
Since the cells are grown on gridded sapphire discs or gridded coverslips, and the gridded pattern is retained throughout the EM preparation, the position of the cell of interest can easily be retrieved for EM acquisition.
Volume Multimodal Imaging – Xray-based targeting (CXEM)
In the EMCF, we employ multimodal imaging in order to identify a target region within a large sample for further structural 3D analysis, by means of TEM-tomography or FIB-SEM.
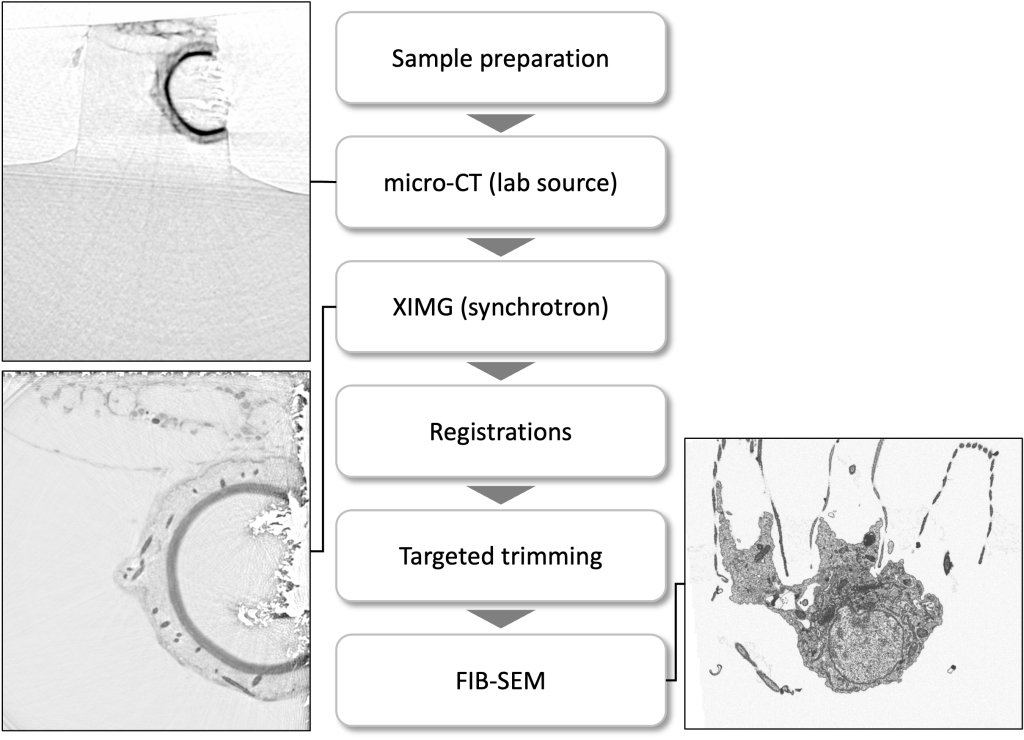
A precise targeting in a 3D volume is feasible when combined with the 3D acquisitions coming from the fluorescent light microscope, a micro-CT scan or synchrotron-based X-ray scanned data (Duke and Schneider labs, EMBL Hamburg).
Below depicts the Volume EM data collection workflow of a specific cell-cell interaction in a marine sponge.
References:
Musser JM, Schippers KJ, Nickel M, Mizzon G, Kohn AB, Pape C, Ronchi P, Papadopoulos N, Tarashansky AJ, Hammel JU, Wolf F, Liang C, Hernández-Plaza A, Cantalapiedra CP, Achim K, Schieber NL, Pan L, Ruperti F, Francis WR, Vargas S, Kling S, Renkert M, Polikarpov M, Bourenkov G, Feuda R, Gaspar I, Burkhardt P, Wang B, Bork P, Beck M, Schneider TR, Kreshuk A, Wörheide G, Huerta-Cepas J, Schwab Y, Moroz LL, Arendt D., Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science. 2021 Nov 5;374(6568):717-723.
Volume Multimodal Imaging – Fluorescence-based targeting
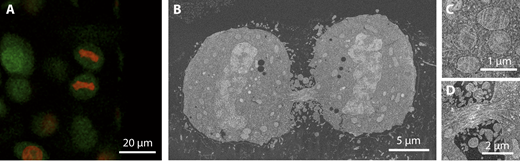
A recent development at the EMCF combines the sample preparation protocols for fluorescence retention and targeted FIBSEM 3D acquisition. The sample block is imaged by confocal fluorescence microscopy after sample preparation. A region of interest is selected based on the fluorescence signal and the ROI labeled by multi-photon laser branding. At the FIBSEM, this region is easily identified and a volume acquired at nanometer scale (down to 5nm isotropic resolution). With this new targeting and acquisition strategy, specific structures in Drosophila, Organoids, HeLa and plankton cells could be resolved.
References:
Ronchi P, Mizzon G, Machado P, D’Imprima E, Best BT, Cassella L, Schnorrenberg S, Montero MG, Jechlinger M, Ephrussi A, Leptin M, Mahamid J, Schwab Y., High-precision targeting workflow for volume electron microscopy. J Cell Biol. 2021 Sep 6;220(9):e202104069.