
Part 4: Topological and structural analysis of proteins
Overview
In addition to analysing the primary amino acid structures of opsins, we can use information from various protein databases to learn more about the molecular structure of the proteins. In this activity, we will use the bovine rhodopsin which we initially identified in Part 1 as example and search the Uniprot database for information on secondary structure and membrane topology (i.e., how the protein structure is positioned with respect to the cell’s membrane). We will then access the Protein Data Bank in Europe (PDBe) to visualise the three-dimensional structure of the bovine rhodopsin.
Your task
Proceed as described below:
1. Search for the bovine rhodopsin in the Uniprot Protein Knowledgebase database to establish the protein’s secondary structure and topology.
2. Discover the three-dimensional structure of the bovine rhodopsin using the Protein Data Bank in Europe (PDBe).
3. Try to answer some of the task questions.
Uniprot and PDBe
1. Open Uniprot in a separate browser window.
2. Search the Uniprot “Protein Knowledgebase (UniProtKB)” by entering the Uniprot accession number (or name) of the bovine rhodopsin in the query box and hitting “search”: Uniprot name: OPSD_BOVIN, accession number: P02699
On the results page, you will find a summary of all scientific information available about this bovine rhodopsin. Scroll through the information and try to answer some of the task questions.
3. To access PDBe (Protein Data Bank in Europe), scroll down to the “Sequences” field and click on “UniParc” next to the accession number. This takes you to a sequence archive which contains links to a diverse range of database entries specific to the protein in question. Identify any links relating to PDB (cf. “Database” column) and, if entries exist, click on the link of the newest entry (the 6th column shows the “last seen”).
4. You have now been forwarded to the bovine rhodopsin entry in PDBe.
5. Click on “View in 3D” and then select the “Astex Viewer”. Please note that you might be asked to grant permission to Java again at this stage.
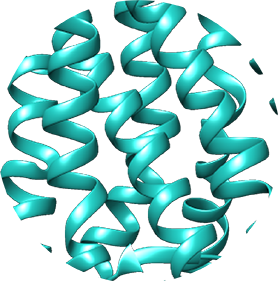
6. You are now viewing a three-dimensional ribbon-cartoon structure of the bovine rhodopsin. You can use your cursor to rotate the structure. Selected features (such as type of structural representation or colour) can be modified either by using the menu on the left or by right-clicking on the image. Have a look at the three-dimensional structure and try to answer some of the task questions.
Questions
1. Looking at the information provided by the Uniprot database, can you answer some of the following questions?
- How many transmembrane domains does the protein have and how are they distributed across the protein?
- Which amino acid residues form the binding site for the chromophore?
- What does the secondary structure of the protein look like (are there more alpha-helices or more beta-strands)?
2. Looking at the three-dimensional structure, can you answer some of the following questions?
- What does the shape of bovine rhodopsin look like?
- Can you identify the individual alpha-helices and beta-sheets? This protein has been crystalised as a dimer, which means that it contains two polypeptide subunits. Can you see the two sets of 7 transmembrane helices?
- Can you localise the chromophore binding site within the ribbon structure?
Activity navigation
Share:
 Čeština
Čeština Français
Français Ελληνικά
Ελληνικά Italiano
Italiano


