
Darwin Tree of Life in 2021: tireless fieldwork and the first beautiful genomes
A look back at some of the 2021 highlights from the Darwin Tree of Life partner institutes

Having overcome a challenging 2020, with plans set back by a global pandemic, the Darwin Tree of Life (DToL) project has flourished in 2021. Teams have been collecting species across Britain and Ireland, from the mountains of Scotland to the sea caves of Wales and the forests of Oxfordshire. Back in the labs, researchers have been processing protists, extracting DNA, and assembling and publishing the first of many high-quality reference genomes.
With 2022 on the horizon, the DToL team has assembled over 200 genomes and published almost 50 of those as Genome Notes – insects, amphibians, fish, echinoderms, one very long worm, and even a wolf. And the project is just starting to ramp up!
Intensive fieldwork, on land and sea
Natural History Museum (NHM), London
This year saw some intensive fieldwork, on land and sea, which kept the NHM DToL team very busy. Fieldwork took place in South-East England and Scotland, with support from NHM curators, NatureScot, Sanger, and a network of experienced entomologists.
In 2021, the team collected around 1,700 specimens of 1,100 species. Highlights were trips to Beinn Eighe National Nature Reserve in the far North-West of Scotland, marine sampling at Millport, and summer days in South-East England’s chalk grassland. The sampling team joined a Dipterists’ Forum trip to Cornwall in August, resulting in a flood of flies for DToL.

Keen folk at NHM provided a steady flow of new centipedes, millipedes, moths, beetles, and wasps for the project. Along with the common and the rare, numerous invasive or newly arrived species have also been sampled to provide a good snapshot of the ever-changing UK fauna.
NHM also had their first Genome Note published – St Mark’s fly (Bibio marci), the team’s first species to go from collection to a whole genome freely accessible to the general public.

An ecological ascent of Ben Nevis
Royal Botanic Garden Edinburgh
The Royal Botanic Garden Edinburgh (RBGE) team spent a total of 63 days in the field from May to October, with sampling events primarily concentrated on the species priority list, focusing on family representatives or target species. An RBGE-led expedition to Ben Nevis resulted in the collection of the ecologically important snowbed species Polytrichastrum sexangulare, the Northern Haircap, only found near the summits of Edinburgh’s highest mountains where small patches of snow persist nearly all year round.

Further sampling highlights include: Eriocaulon aquaticum, the pipewort, restricted to a few sites on the Scottish Isles, collected from Skye; Scheuchzeria palustris, aptly named the Rannoch Rush as it only occurs on or close to Rannoch Moor; and Diapensia lapponica, the pincushion plant, found on only one mountain top at Glenfinnan in the Highlands. Each of these three species is the only representative of their family found in the UK.
Overall, in the main collecting season from May to October, the RBGE Genome Acquisition Laboratory (GAL) collected 2,091 DToL samples, representing 143 species of vascular plants, 84 bryophytes, and 3 lichens. An additional 149 samples, representing 25 plant species, have come through the RBGE GAL in collaboration with Buxton Climate Change Impacts lab. Ultimately, 1,716 samples were shipped to the Sanger Institute, including 144 bryophyte samples for the plant research and development panel and 333 samples for the Apple Day project. In total, RBGE has sampled 389 plant species to November 2021 (185 bryophyte species from 79 families, and 204 vascular plant species from 87 families).

Shock sighting of a silver fly – the first in Wytham Woods
University of Oxford, Wytham Woods
After a couple of years intensively sampling Wytham Woods, this team was not expecting to find many more species that were ‘new for the site’ this year – how wrong they were! In fact, 2021 proved to be a bumper year for rare and interesting species. And one discovery stood out from the rest.
On 8 July, the team undertook a day of intensive sampling in an area of the woods known as ‘the Dell’. They were mainly targeting beetles that live in dead wood as there were several families in this group that were yet to be collected. Liam Crowley, DToL Postdoctoral Researcher, spent some time examining a particularly huge veteran beech which had fallen over, revelling in the diversity of saproxylic insects it hosted.
Whilst Liam was admiring the abundance of solitary wasps buzzing in and out of their nest holes in the tree, a large fly flew over his shoulder and landed on the trunk right in front of him. ‘Therevid!’ he thought, great, they hadn’t collected that family yet!

As Liam moved nearer, net in hand, and got a closer look, he nearly fell over with shock. The fly was bright silver! The common woodland species in this family are all brown. Silver species are usually only found on coastal dunes or are vanishingly rare.
Sure enough, he was able to confirm this specimen as Pandivirilia melaleuca, the forest silver-stiletto fly. This species is very scarcely seen and was previously only known from Windsor and the surrounding area, as well as a second cluster in West Gloucestershire and South Worcestershire. Wytham Woods falls neatly in between these two groupings. Not much is known about the biology of this species, but the larvae are believed to develop in the heartwood of dead trees where they prey on saproxylic beetles. This specimen represents the first species from the family Therevidae submitted for full genome sequencing.

Journey to a seldom-studied protist paradise
University of Oxford, Protist Group
Priest Pot is a body of freshwater, about one hectare in surface area, near the village of Hawkshead in the Lake District. The site, part of a National Nature Reserve, appears fairly unremarkable. However, these waters are teeming with a huge variety of single-celled organisms – known as protists – blooming and feeding in their own complex ecosystem beneath the surface.
The DToL team scouted out this seldom-studied protist paradise in 2020, finding the site surrounded by swampy undergrowth and a barbed wire fence. Getting in touch with the landowner took persistence. When Estelle Kilias’s emails went unanswered, she even tried sending a letter. An email response soon followed, and progressed to many friendly phone calls.
It was September 2021 by the time the Oxford University team were out on Priest Pot’s waters. “You feel every bone,” Estelle recalls of the fieldwork. First, they lowered probes to find the most interesting environmental conditions in the lake – these determine whether rarer protists will be thriving. Next, they plunged in five-litre Niskin bottles to precisely collect the water they needed, pouring the samples into 20-litre carboys to take back to shore. Ultimately, a hundred litres of sample water were collected and taken back to the lab in Oxford, where their protist treasures are being painstakingly discovered.

Seeking genomes beneath the surface
It’s been a busy year of collecting and processing for the Marine Biological Association DToL team, with 568 different species collected from 351 different families and 188 different orders. That includes marine lichen, fungi, algae, fish, crabs and more. Although there have been quite a few field trips, a huge amount of these species have been collected right on the MBA’s doorstep in Plymouth.

Britain and Ireland has the most incredible diversity of marine life – this is, after all, an archipelago – but a lot of it is hidden to most of the people who live here. Team MBA has set out to uncover what lives beneath the waves: they have been out on boats, diving through marinas, wading through rockpools, clambering over muddy tidal flats and scrambling into watery caves to find all sorts of creatures living in all sorts of strange places.
Below is a selection of some of the amazing marine life they found this year.

1. The parchment worm (Chaetopterus variopedatus) is a bioluminescent polychaete worm that builds itself a papery tube from secreted mucus.
2. Here is the black lichen Lichina pygmaea. Lichens are a stable symbiotic relationship between algae or cyanobacteria and fungi. So little is known about lichens that DToL teamed up with other MBA colleagues working on lichens to help find and ID these tricky species. Lichens are tiny ecosystems and cleaning them up to prepare them for DNA barcoding was a long and arduous task, but MBA’s in-house barcoder Joanna Harley did a marvelous job, getting good reads for many of these notoriously difficult-to-isolate organisms.
3. This is a sea spider, Pycnogonum litorale, one of 1,300 species world-wide. The scientific order they belong to is called Pantapoda, meaning ‘all feet’, very appropriate for these leggy animals.
4. This beauty is a worm pipefish (Nerophis lumbriciformis). These animals live mostly in and around rockpools. Like other pipefish and seahorses, the males carry the young and they mate for life, so if you see one, make sure to leave it be!
From big data to beautiful genomes
Wellcome Sanger Institute’s Tree of Life programme
To date, well over 200 DToL genomes have been assembled and curated by the dedicated teams of bioinformaticians at Sanger – the vast majority of them in 2021.
The Tree of Life Assembly (ToLA) team and Genome Reference Informatics Team (GRIT) are responsible for turning masses of DNA data – those A, C, G, and T base pairs – into beautifully assembled and curated genomes. Crucially, these are arranged in chromosomes, to reflect biological reality as closely as possible, before being released to the scientific community.
“We have received triple the number of curation requests compared to 2020,” explains Jo Wood, who heads GRIT. “As we do this, to meet the ambitious goals of the project, we are constantly looking for ways to increase throughput and improve turnaround whilst maintaining quality. A huge amount of effort has gone into streamlining, automating, and generally reducing the human hands-on time required.”
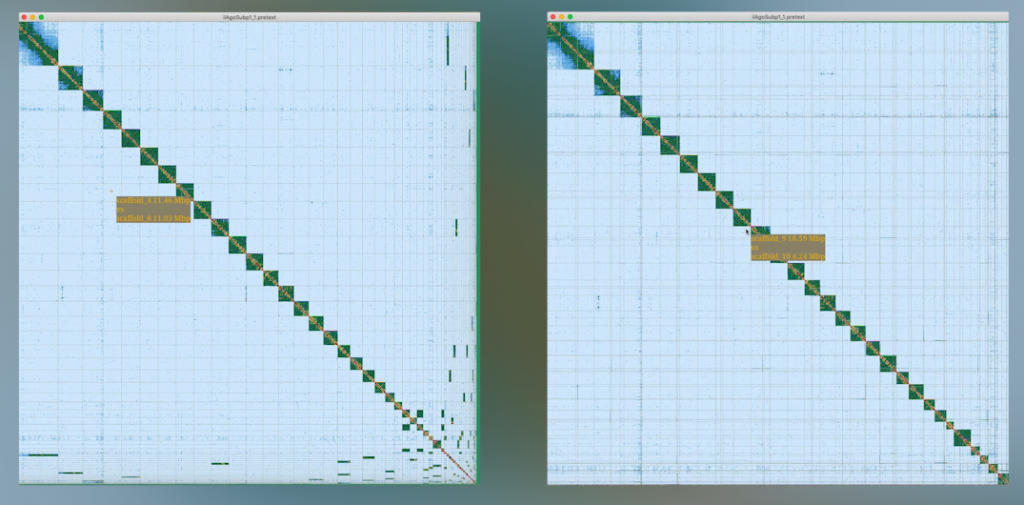
The two teams work closely together to make sure the final genomes are of the highest possible quality. For example, this year GRIT spotted some missing data when curating an apple genome. The ToLA team then investigated and uncovered a couple of bugs in the programs.
“Our pipeline is working on a huge variety of different organisms, such as plants, worms and lepidoptera,” says Marcela Uliano-Silva, a senior bioinformatician on the ToLA team who has also written a tool for assembling mitochondrial DNA from Pacbio HiFi reads this year. “To put into perspective how far the science has come: the human genome was published two decades ago, having taken 13 years, almost $3 billion, and nearly 3,000 scientists. We’re now producing several new genomes per week, in much higher quality and at the chromosomal level.”
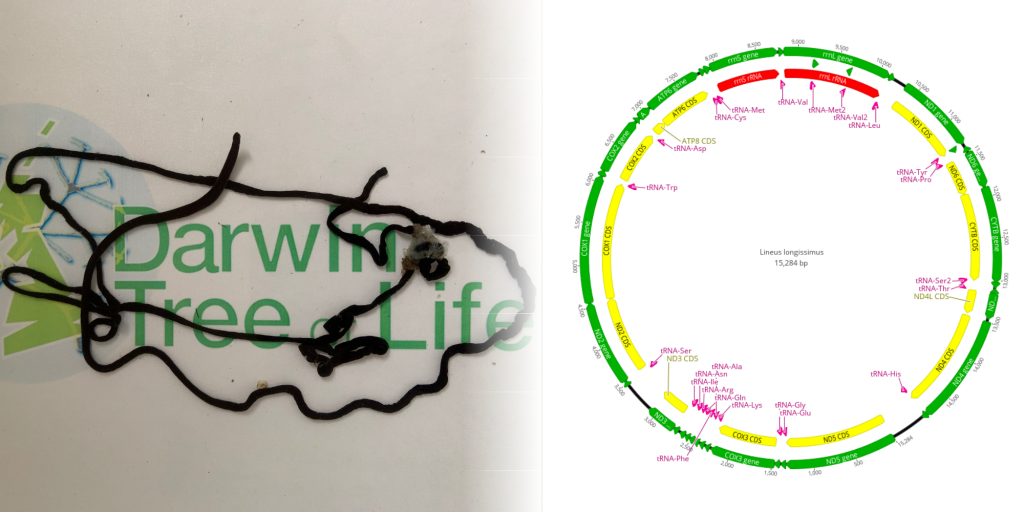
One of the genomes the Sanger team worked on was that of the super-stretchy ribbon worm, Lineus longissimus – which, at full length, is the longest animal in these islands. Its genome, however, is just an eighth the size of the human genome. Compare that to the mistletoe genome which is 30 times larger than the human genome – and is one of the trickier genomes the team expects to sequence and publish in 2022.
Open access data, phylogeny, and Our Animal DNA
EMBL’s European Bioinformatics Institute (EMBL-EBI)
This year has been all about butterflies and moths for the DToL team at EMBL-EBI. The Ensembl Compara team generated a cactus alignment – a method for creating multiple genome alignments – of around 90 butterfly and moth (Lepidoptera) species along with several closely related species.
Open access to the annotated genome assemblies created will enable more detailed exploration of the evolution of genome structure within the Lepidoptera. These genomes are freely available to anyone through Ensembl Genomes and the DToL Data Portal.
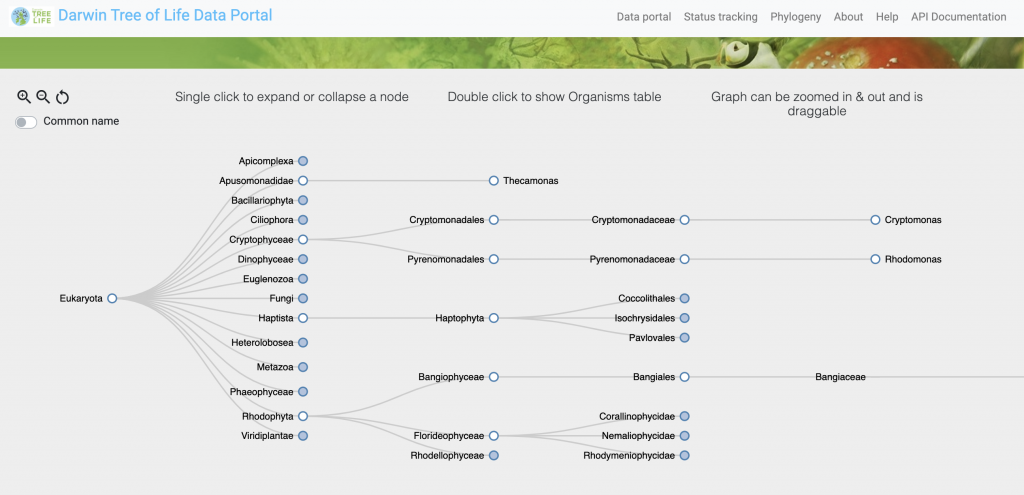
The EMBL-EBI team also updated the DToL Data Portal to include a phylogeny browser that allows users to navigate a tree-like structure and see what data is available for a particular clade, family, or genus.
And in public engagement news, EMBL-EBI now offers a unique DToL public engagement activity – Our Animal DNA, developed by Ensembl Outreach. This activity is available for school students aged 16–18 years and will introduce them to bioinformatics tools and techniques through an introductory video from EMBL-EBI scientists, in addition to online classroom activities.
Barcoding the Broads
In September, the Earlham Institute launched the first in-person training workshops for its DToL public engagement programme: Barcoding the Broads. The sessions are tailored for a non-specialist audience and focus on a technique called DNA barcoding, where an organism can be identified by analysing unique patterns of DNA within its genome.
The methods are straightforward and reliable, thanks to huge advances in sequencing technology and the miniaturisation of lab equipment, allowing anyone to identify an organism with a little bit of training and support.
The team at the Earlham Institute, led by Sam Rowe, have had some fantastic initial feedback from teachers, naturalists, sixth form students, and education professionals who attended the first few sessions. Plans are also in place for future work to help communities in Norfolk explore the biodiversity on their doorstep.

By the end of 2021, training would have been provided to 50 people with almost 100 more hoping to attend a workshop in early 2022. In partnership with Kew Gardens, the team was also successful in an application to the Enabling Connections Fund, which will allow them to embark on an exciting new collaboration with the Norfolk Fungus Study Group to explore DNA barcoding with fungi on the Norfolk Broads.
DNA barcoding workshops run from 9:30 a.m. to 4:00 p.m. in the Norwich Research Park and are free to attend for groups of up to nine people. If you would like to get involved to learn new techniques for your research and/or education work then please visit the Earlham Institute website and get in touch with the team.

This article was originally published on the DToL blog where you can find the latest news and updates from across the project.


