
Come together
Thanks to a technique developed by their lab a few years ago, scientists at EMBL Grenoble have the first experimental proof that a key cellular machine forms by uniting pre-assembled modules.
“Currently, we can get a pretty good picture of what complexes exist in a cell, but what is uncharted territory is how the cell actually assembles these complexes,” says Imre Berger from EMBL Grenoble.
The complexes Berger is referring to are large cellular machines, built up from many different building blocks. Such machines carry out most of the fundamental tasks in the cell, notably fine-tuning the readout of our genes.
A case in point is a complex Berger has been studying for a number of years: TFIID (pronounced tee-eff-two-dee). “Virtually all protein-coding genes in human cells are controlled by TFIID – it is a central regulator of human gene transcription, and we still do not really know how this essential complex functions,” he says.
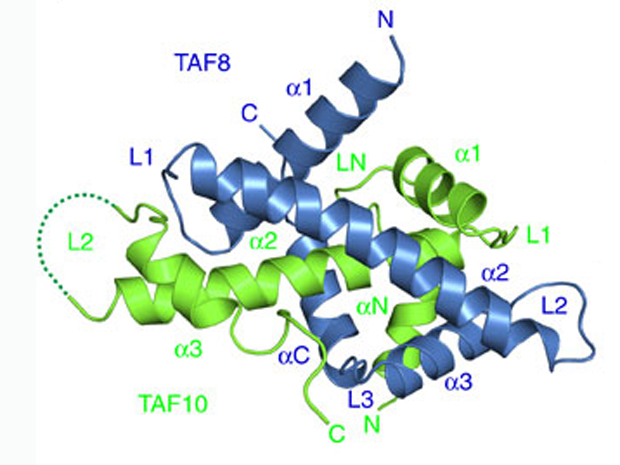
Thanks to techniques previously developed in the Berger lab, the EMBL scientists were able to create a probe to look for one particular piece of TFIID, called TAF2, inside cells. Working with Lazlo Tora’s lab at IGBMC in Strasbourg, they found that TAF2 links up to two other building blocks – TAF8 and TAF10 – in the cytoplasm, forming a stable module which then travels into the nucleus to bind with the ‘core’ of TFIID – whose structure Berger’s lab revealed in 2013 – to form the complete complex. What’s more, they discovered that this module has to form in order for the three building blocks to even make it into the nucleus: when the researchers genetically engineered cells to produce no TAF8, TAF2 and TAF10 became ‘trapped’ in the cytoplasm.
This is the first time that scientists have been able to show through experiments that TFIID isn’t built in a single step, with all its different pieces coming together at once in the nucleus, but rather that it is assembled from pre-formed modules.
I feel super-excited because it’s a new, original angle which one can experimentally address
The findings support the idea that answering the question of how the cell goes about building a complex can help to explain how it works. The TAF2-TAF8-TAF10 module could play a role of its own, before it is even ‘hooked up’ to the TFIID core. In some tissues – like the ovaries and sperm cells – TFIID is missing certain building blocks, and there is at least one other complex – SAGA – which uses some of the same building blocks as TFIID. So by choosing which modules to include, or which complex to allocate a module to, the cell could be controlling which genes are acted upon at a given time – for example, to turn on specific genes needed to produce and maintain a sperm cell.
Berger and colleagues would now like to look at other pieces of TFIID, to see if and where they also assemble into ‘stand-alone’ modules, and investigate at what stages different modules are built and brought together. In the longer run, they’d also like to compare the different tissue-specific versions of TFIID, and study how a cell ‘decides’ whether to use available building blocks to produce TFIID or SAGA and how this affects gene activity.
“I feel super-excited about this, because it’s a new, original angle which one can experimentally address,” Berger says, “and we are in a particularly privileged position to do so, because we can make all these pieces with the quality you need to generate really specific probes.”


