Limblab: pipeline for 3D analysis and visualisation of limb bud gene expression
BMC Bioinformatics 12 January 2026
https://doi.org/10.1186/s12859-025-06264-4
Researchers at EMBL Barcelona have developed an open-source tool that makes working with complicated volumetric imaging data easier
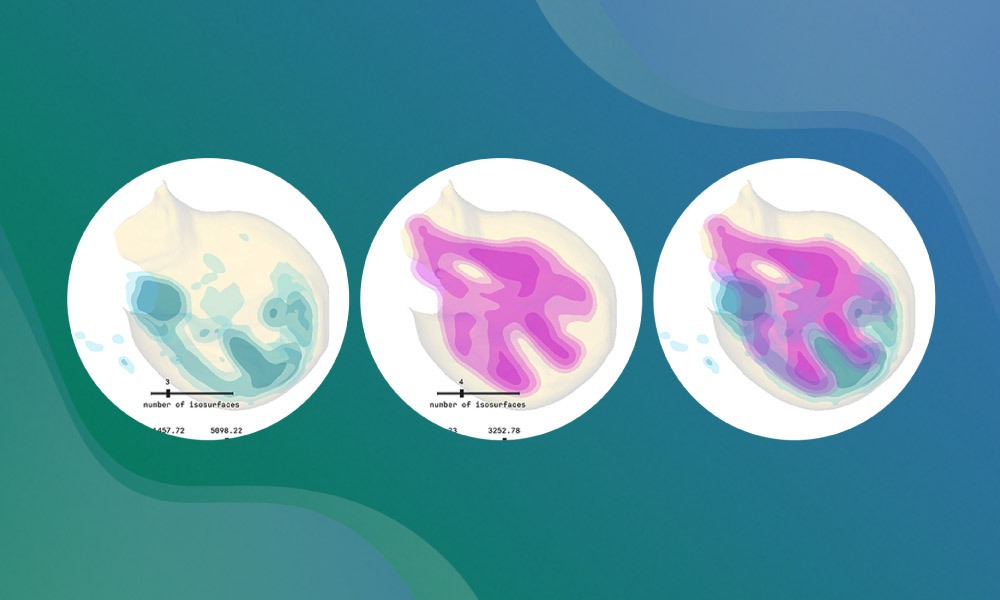
Studying the shape of tissues and organs is critical to understanding how they are formed. Embryonic development happens in three dimensions, but many studies are limited by the use of two-dimensional approaches and images to describe three-dimensional processes. To overcome this challenge, researchers at EMBL Barcelona have created LimbLab – an open-source pipeline made for three-dimensional visualisation and analysis of growing limb buds.
The platform was primarily designed to study mouse limb development, but the concept can be useful for any researcher working with complicated volumetric imaging data. The researchers describe the platform in a new study published in the journal BMC Bioinformatics.
“We developed LimbLab because we realised that current tools miss crucial aspects of embryonic development and are not designed specifically for developmental biology,” said Laura Aviñó-Esteban, first author of the work and PhD student in EMBL’s Sharpe Group.

Researchers studying limb development need specific software tools for their work. For instance, researchers may need to assign a developmental age to certain samples based on their visible features, or they might have to align or morph images of samples to allow accurate and consistent comparisons.
These tools exist in 2D, like eMOSS and LimbNET, but not in 3D. LimbLab bridges this gap by enabling 3D visualisation of gene expression patterns. It achieves this through a modular workflow in Vedo, an open-source Python library developed at the Sharpe Lab, which enables seamless 3D analysis and high-fidelity and aesthetic rendering of meshes and volumes.
First, the pipeline cleans the raw volumetric data obtained from a microscope, removing noise and artefacts. Then it extracts information about tissue surfaces to build computational structures called ‘meshes’, which are computationally efficient to work with. After this, the pipeline analyses the sample to give it a developmental age and aligns or morphs the sample with a reference model. LimbLab also provides advanced visualisation methods that help researchers to explore and present gene expression in full 3D. Each step is logged and standardised, which improves reproducibility.
The importance of this work goes beyond limb development. Many areas of biological research, like those using organoids, tumours, or engineered tissues, depend on 3D volumetric imaging. LimbLab shows how a specialised pipeline can organise messy 3D data and make it clear and aligned. It also underscores the importance of reproducibility in imaging research, where small differences in processing can result in very different biological interpretations. LimbLab is also a proof-of-concept: it shows how special computational tools can change the way we analyse 3D biological data.
LimbLab is not only a technical improvement, it is a step towards making developmental biology research more quantitative, reproducible, and accessible. The pipeline is open source, easy to install, and has full documentation. While the present version is optimised for mouse limb buds, the researchers plan to adapt it for other species and tissues to help answer bigger questions about regeneration and evolution.
BMC Bioinformatics 12 January 2026
https://doi.org/10.1186/s12859-025-06264-4