
AlphaFold: a game-changer for structural biology
EMBL-EBI teams up with DeepMind to make breakthrough AI-powered protein structure predictions freely available to the world.
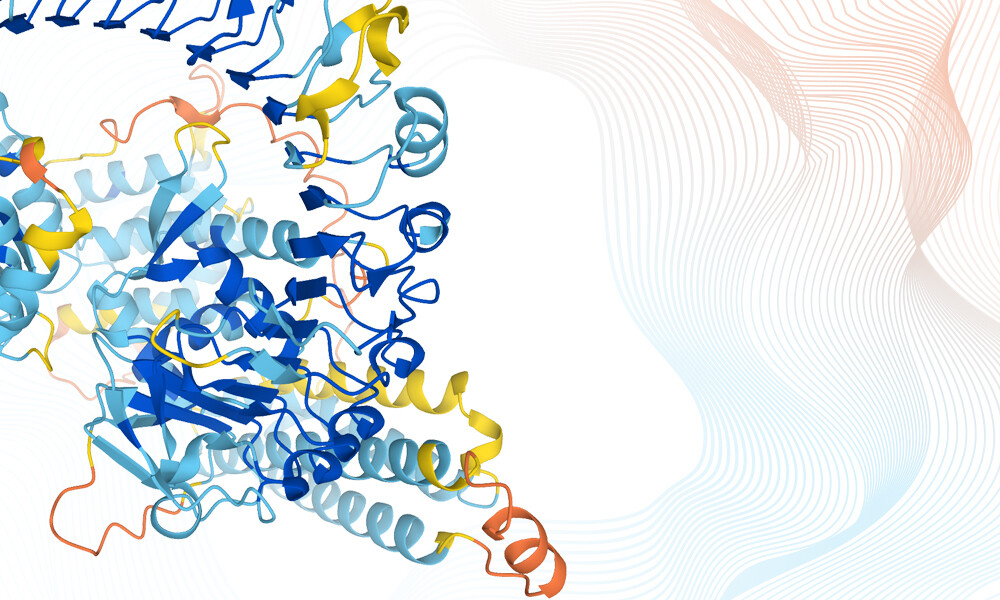
Proteins play many roles in the biological world. They transport nutrients, trigger chemical reactions, and build the structures that make up all living things. To fully understand what a protein does, scientists need to know its 3D structure, and this is no easy task.
Since the 1950s, scientists have used a range of methods to uncover hundreds of thousands of protein structures and made the data accessible through public archives. Building on these data, DeepMind developed AlphaFold – an AI-powered system that can accurately predict millions of protein structures.
In 2021, EMBL-EBI and DeepMind entered into a collaboration to develop and release the AlphaFold Protein Structure Database – an open platform where anyone can search, analyse, and download AlphaFold predictions.
The beauty of AlphaFold is that it was trained using data from public resources – including the Protein Data Bank (PDB), UniProt, and MGnify, which are all co-hosted at EMBL-EBI. Building on EMBL-EBI’s decades of data management expertise, DeepMind was able to quickly provide public access to AlphaFold predictions and crosslink them to existing data resources, making it much easier to discover them. DeepMind and EMBL-EBI are working to extend the database to over 100 million predictions.
Many labs at EMBL and around the world have used the data to gain new insights into protein science:
- Jan Kosinski at EMBL Hamburg used AlphaFold and electron cryo-microscopy data to build a new model of the nuclear pore complex, exemplifying how AlphaFold can support experimental structural biology to understand subcellular architecture.
- Pedro Beltrao at EMBL-EBI and collaborators used AlphaFold to predict complex protein interactions, expanding understanding of human cell biology.
- Jan Korbel at EMBL Heidelberg created a de novo protein design framework based on AlphaFold, which could help design new proteins for applications such as processing waste or degrading plastics.

“I love that the collaboration between DeepMind and EMBL will make all the knowledge about protein structure open to all.”
— Jacques Dubochet, Nobel Laureate for Chemistry 2017; Group Leader at EMBL, 1978–1987

“The EMBL-EBI and DeepMind collaboration that created the AlphaFold database is a big win for open science. Life sciences research will be able to make quantum jumps with this publicly accessible data.”
— Mihaly Varadi, Project Leader, Protein Data Bank Europe and AlphaFold Protein Structure Database, EMBL-EBI

“Tools like AlphaFold require dedicated computing infrastructure to run, which takes a whole team to manage. I look forward to seeing how AlphaFold will give us new insights, e.g. into how neural networks can be applied to turbulence modelling – another large grey area on the science map.”
— Jurij Pečar, HPC Engineer, IT services at EMBL Heidelberg
Explore more 2021 EMBL structural database highlights:
References
Mosalaganti S et al. (2021). Artificial intelligence reveals nuclear pore complexity. bioRxiv, 2 November 2021. DOI: 10.1101/2021.10.26.465776
Burke DF et al. (2021). Towards a structurally resolved human protein interaction network. bioRxiv, 9 November 2021. DOI: 10.1101/2021.11.08.467664
Jendrusch M et al. (2021). AlphaDesign: A de novo protein design framework based on AlphaFold. bioRxiv, 12 October 2021. DOI: 10.1101/2021.10.11.463937

