
Read the latest Issue
EMBL’s new group leader based in Hamburg aims to create models of infection cycles
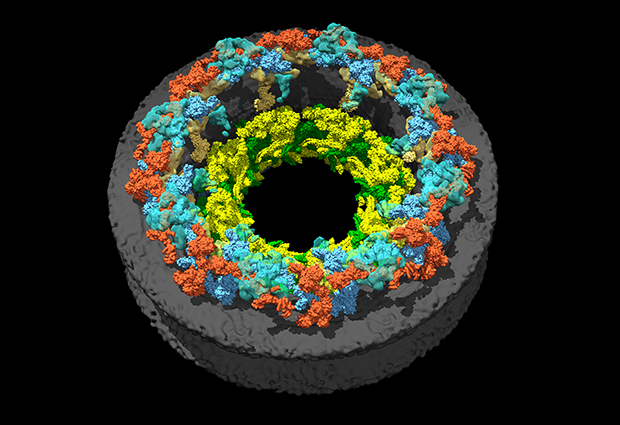
Working as part of the Centre for Structural and Systems Biology (CSSB) in Hamburg, new EMBL group leader Jan Kosinski will lead a team of structural and systems biologists to model structures and processes.
My work has been predominantly in structural biology and bioinformatics: I use a computer to predict what the structure of proteins and protein complexes looks like based on theories or data. For instance, during my postdoc at EMBL in Heidelberg I studied the nuclear pore complex, a passage into a eukaryotic cell’s nucleus that plays a crucial role in controlling which molecules enter or exit this part of the cell. Our team was interested in what the structure of the channel looks like so that we could understand how transport between the nucleus and the cytoplasm works. But the nuclear pore complex is probably the biggest complex in the eukaryotic cell, and we are not able to determine its structure using a single method such as X-ray crystallography or cryo-electron microscopy. So, together with colleagues, we collected different types of data from different types of experiments and developed computational methods to integrate this data and create a model of the human nuclear pore complex.
In my new lab in Hamburg, we continue to integrate models and data in order to understand tricky structures. But we’re also embarking on a completely new direction: the integrated modelling of the viral infection cycles. Many viruses need to pass through the nuclear pore complex in order to enter the nucleus of the cell. To do this they may pass through it by tricking the cell’s machinery. Some viruses literally cut the pore into pieces and strip off proteins to block the cell exporting key genetic information, hindering their defense mechanisms or reprogramming the pores so that they transport molecules that support viral replication.
I’d like to get to the stage where we can create computational models of entire diseases or infection cycles
My ultimate goal is to understand which viral proteins interact with which host proteins and what kind of mechanisms they use to hijack the host’s cells. To do this, we need to collect data about interactions that happen during the infection, protein expression and other changes that we can measure, and then use this information to reconstruct the subsequent stages of the infection cycle. Since the data collected during the interactions is usually very noisy, we also want to develop new methods to sieve real interactions from the background noise. My immediate challenge is to build a team with the skills necessary to achieve this.
Excited, for sure! First off, I just need to organise myself and find my way. There are some things I will certainly miss from being a postdoc – the times when I could plan several days simply to work on developing an algorithm without distraction! But the prospect of driving new projects and leading and developing my team is something I am incredibly excited about. I love the EMBL culture and how it works. CSSB is a really unique place, especially if you want to collaborate with other scientists and institutions; the setup really promotes this kind of work and I have already started discussing collaborations with several researchers. Like in Heidelberg, everyone in Hamburg is really supportive, helpful and very friendly and it is great to be here.
My mother: although she is not a scientist, she has always been curious about life. We always had lots of biology books on our shelves at home. The conversations we had stuck with me and eventually I decided I wanted to become a scientist. I am Polish, and one day I saw an article in the Polish edition of Scientific American about some Polish researchers who had managed to model proteins and were very good at it. When I went to study in Warsaw I remembered them, tracked them down and ended up working in their lab – I was hooked!
I’d like to get to the stage where we can create computational models of entire diseases or infection cycles. To expand from atoms to cells – or even tissues and organisms – and use these models to predict outcomes and guide our experiments. For instance, what happens if you interfere with a particular protein? How does it affect the infection? We are only at the very beginning of being able to do this, and I am very motivated to take the steps needed to make this a reality.
Keep going! If something is hard, give it some extra time and effort. Keep pushing, be persistent and sooner or later you will achieve your goal.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive