Read the latest Issue
Two in one
Multi-tasking protein provides new approaches for anti-tuberculosis drugs
In a paper published online today in PNAS, scientists from the European Molecular Biology Laboratory (EMBL) in Hamburg, Germany, reveal new insights into the workings of enzymes from a group of bacteria including Mycobacterium tuberculosis, the bacterium that causes tuberculosis. The new findings present possible new opportunities for developing organism-specific drugs, which target the pathogen but leave other microorganisms, which are beneficial to us, untouched.
Tuberculosis remains one of the largest threats to human health worldwide, and one of the most frequent causes of death in HIV patients. With the increasing emergence of strains of Mycobacterium tuberculosis that are hyper-resistant to drugs, it becomes ever more urgent that novel treatments be developed, and the search for novel strategies for drug development is an important step in this process.
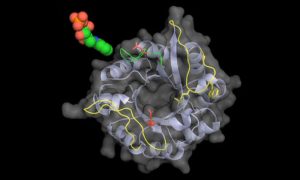
In the current study, Matthias Wilmanns and his group at EMBL identified a multi-tasking enzyme from Mycobacterium tuberculosis that catalyses reactions on two different molecules, or substrates. In most organisms, cells need two specific enzymes, known as HisA and TrpF, in order to produce two essential amino acids – histidine and tryptophan. However, in Mycobacterium tuberculosis, the encoding gene for TrpF is missing, and the two reactions are instead catalysed by a single enzyme, which is able to recognize and bind to two different substrates. This enzyme is known as PriA.
Using the Mycobacterium tuberculosis version of the PriA enzyme as a model, the researchers were able to unravel the hitherto unknown mechanism of bi-substrate specific binding observed in this group of bacteria.
“When we solved the three-dimensional structure of PriA, we found that it has the unique ability to form two different substrate-specific active sites,” Wilmanns says: “it can form a reaction-specific active site, or undergo what we call ‘substrate-induced metamorphosis’ to form a different active site.”
To further verify these observations, Wilmanns and colleagues screened 20,000 small molecule compounds, and identified a handful which inhibited both PriA-catalysed reactions but had no effect on TrpF activity.
“We believe that this ability for bi-substrate catalysis in Mycobacterium tuberculosis could be a new opportunity for future drug development,” Wilmanns concludes: “This organism-specific reaction process could be exploited, since only the pathogen but none of the other bacteria living in or on humans, many of which are important for our well being, would be targeted.”