Thomas Quail
Group Leader
ORCID: 0000-0002-3569-0318
EditPrinciples of genome self-organisation

Group Leader
ORCID: 0000-0002-3569-0318
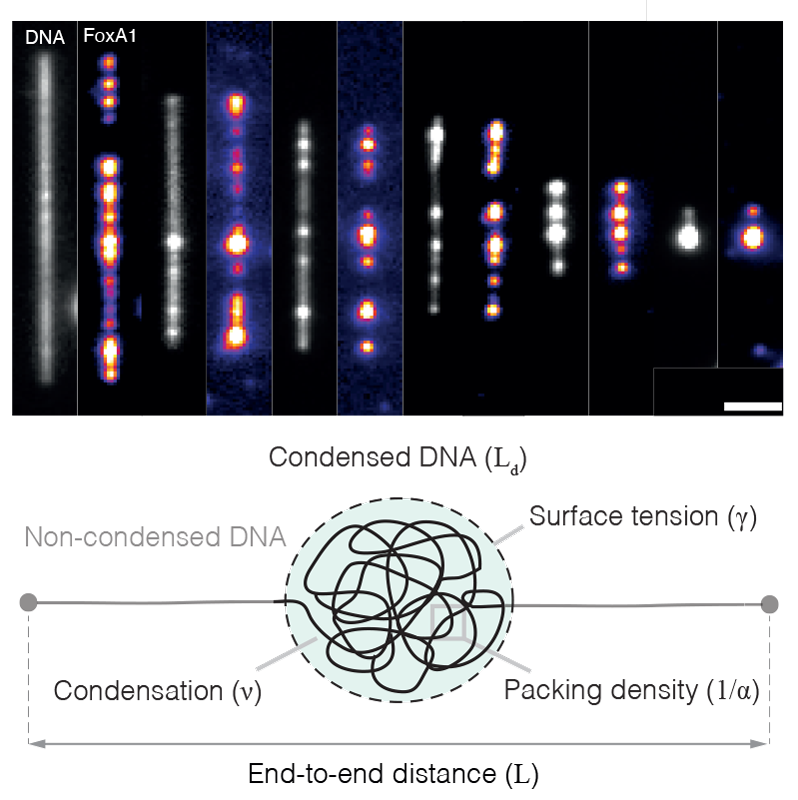
EditHow the cell organises its contents represents a remarkable example of collective behaviour in which thousands of factors — such as proteins, nucleic acids, and lipids — self-organise to compartmentalise biochemical reactions, build intracellular structures, and perform mechanical work. One striking example of collective behaviour in the cell is the organisation of the genome. In every eukaryotic cell, two meters of DNA are packaged into the cell nucleus, a subcellular organelle with a diameter of roughly 5-10 μm. Proteins drive this process, generating forces that compartmentalise the genome into nucleosomal arrays, topologically associated domains, chromatin domains, and chromosomal territories. Genetic, proteomic, and structural studies have revealed the identity of many of these force-generating proteins, and techniques such as chromosome conformation capture have generated static pictures that have demonstrated the genome’s non-random organisation. However, how these proteins work collectively together to coordinate this organisation remains unclear and raises many questions. What physical mechanisms drive genome organisation? How does the biochemistry of individual factors regulate this organisation? How do these physical mechanisms regulate processes in the nucleus such as transcription, DNA damage repair, and DNA replication?
To address these questions, we adopt a broad, interdisciplinary, and quantitative approach, combining in vitro biochemistry, quantitative microscopy, single-molecule biophysics, Xenopus egg extracts, and theory. We focus on how the condensation of proteins can generate forces that bridge distally located DNA (Quail et al., 2021) and how these condensates compete with energy-consuming motors that drive genome organisation (Golfier et al., 2020). To make progress on these problems, we directly image how proteins drive genome organisation at different length scales (from single molecules to intact nuclei), extract quantitative data from these experiments, and then develop theoretical models that generate testable experimental predictions.
We have just started understanding how proteins drive genome self-organisation across different length scales and have many open research goals and directions.
We are an inclusive and curiosity-driven lab, tackling the problem of genome self-organisation using a wide range of backgrounds, tools, and expertise that span biochemistry, cell biology, and physics. We foster an open lab environment focused on scientific exploration and growth. New ideas, approaches, model systems, and interdisciplinarity in any form is encouraged — reach out if you are interested in joining!