Edith Heard
Group Leader (Incoming/Outgoing)
ORCID: 0000-0001-8052-7117
EditEpigenetic mechanisms in development and disease

Group Leader (Incoming/Outgoing)
ORCID: 0000-0001-8052-7117
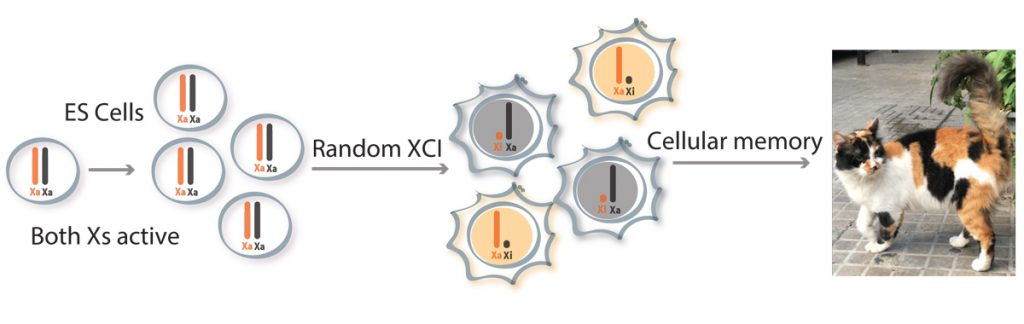
EditOur laboratory is interested in epigenetic mechanisms of mammalian development and disease. A powerful example of epigenetics in mammals is X-chromosome inactivation (XCI), the process whereby one of the two X chromosomes in females becomes transcriptionally silenced during early development. This enables dosage compensation for X-linked gene products between XX females and XY males. Our group studies XCI in order to gain insights into the mechanisms of initiation and maintenance of differential gene expression states and to explore the importance of gene dosage. We use a combination of molecular genetics, cell biology, genomics and single-cell approaches on embryos and cultured cells as well as in tissues and tumours. We study the roles that non-coding RNA, chromatin changes and chromosome organisation play in XCI and other epigenetic processes.
Our lab has had a long-term interest in the X-inactivation centre (Xic) locus that triggers XCI, through the monoallelic up-regulation of the long non-coding RNA (lncRNA) Xist. Our work in early embryos revealed that XCI is remarkably dynamic during early embryogenesis and that different mammals show different modes of initiating XCI. We have defined some of the earliest chromatin changes implicated in XCI and have also shown that in breast cancer, Xi chromatin organisation is disrupted. In another project, we identified a number of autosomal loci showing monoallelic expression that is tissue-specific and variable. We found that many of these loci are also involved in autosomal dominant disorders.
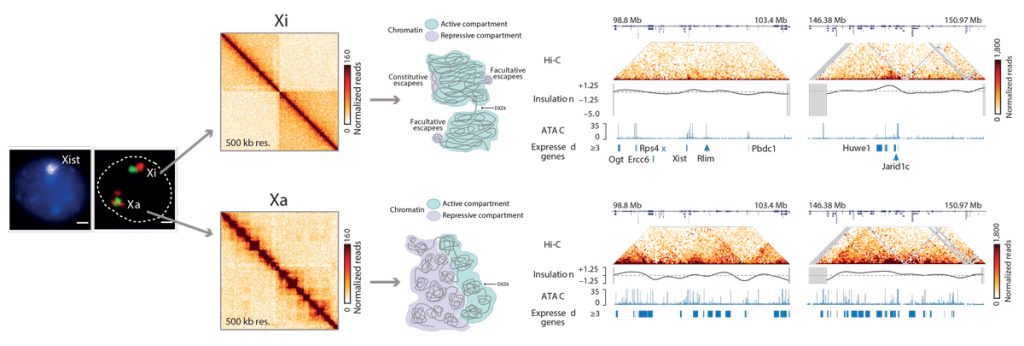
Through our work on the X chromosome, our laboratory has also contributed to a deeper understanding of the role of chromosome architecture in gene regulation. In collaboration with Job Dekker, our recent work on the Xic uncovered the existence of topologically associating domains (TADs) using a chromosome conformation capture approach. We found that spatial partitioning of the Xic into TADs seems to be important for the appropriate developmental regulation of Xist. Using HiC sequencing techniques, we also discovered that the inactive X chromosome is folded into two mega-domains and is globally depleted of TADs, except for regions that escape XCI. Although most genes are silenced on the inactive X, some genes can escape either constitutively or in a variable (facultative) manner and this is linked to local TAD architecture.
We combine genetic engineering and genomics with a range of cell biology and imaging approaches to study:
The Heard group has been supported by two ERC Advanced Investigator Awards, EPIGENETIX and XPRESS. EPIGENETIX (2010-2015) focused on understanding the basis for monoallelic gene expression at the level of the X-inactivation centre, led to the discovery of topologically associating domains (TADs) and identified numerous autosomal loci that also show random monoallelic gene expression. The XPRESS ERC (2016-2021) focused on deciphering the molecular mechanisms underlying gene repression and escape during X-chromosome inactivation. This has led to insights into the roles of early chromatin changes in Xist-mediated silencing as well as the role of the SPEN protein as an integrator of multiple pathways that participate in gene silencing on the X chromosome.