Research
Integrator complex and snRNA 3′-end processing
Integrator complex was discovered as a factor required for the 3′ end processing of metazoan snRNAs, and its coupling to the transcription termination. Recent studies show that the Integrator can destabilize promoter-proximally paused RNAPII, leading to nascent RNA cleavage and transcription attenuation in a wide range of protein-coding genes.
In 2017, we set off to determine the structure of the Integrator complex by cryo-EM. Our initial approach relied on purification of the endogenous complex by affinity chromatography. In order to do that, we generated stable HEK293F cell lines expressing tagged variants of all known Integrator subunits. These cell lines were used to perform tandem affinity purification followed by quantitative mass spectrometry experiments. Based on MS results we identified 3 stable modules of the Integrator complex (INTS5/8, INTS10/13/14 and previously described INTS4/9/11). We performed recombinant expression and purification of all 3 modules in insect cells and characterized them biochemically. We determined cryo-EM structure of the 250kDa INTS4/9/11 complex at overall 3.5Å resolution (Pfleiderer and Galej, Mol Cell 2021). Our structure reveals the architecture of the nuclease heterodimer and together with the supporting biochemical and in vivo reported data provided new insights into the mechanism of nuclease heterodimer assembly and identified a putative RNA binding path within the complex.
Structural studies of the branch site recognition in mammals
Recognition of the intron branch site (BS) by the U2 snRNP is a critical event during spliceosome assembly. In mammalian cells, BS is initially recognised by the SF1 (mBBP) in cooperation with the U2AF2 (U2AF65), which binds the polypyrimidine tract (PPT) sequence. Concomitantly, U1 snRNP binds to the 5’-SS and together with SF1 and U2AF2 forms the first, ATP-independent spliceosome assembly intermediate known as the complex E. The U2 snRNP is loosely associated with complex E and its stable incorporation into the prespliceosome (complex A) requires ATP and the formation of base-pairing interactions between the BS and the U2 snRNA. While parts of the U2 snRNP structure have been determined as a component of yeast and mammalian spliceosomes, there is no high-resolution structural information for the 17S U2 snRNP and early splicing complexes in mammals (i.e. E and A).
In order to bridge this knowledge gap, we established a pipeline for genome engineering using CRISPR/Cas9 approach, and used it to knock-in affinity tags into components of the human spliceosome in the HEK293F cell line. Endogenous tagging of the HTATSF1 allowed us to isolate 17S U2 snRNP, containing U2 snRNA and > 20 protein subunits with the total MW >1MDa. Next, we investigated interactions of this complex with a model RNA substrate as well as its ATP-dependent remodeling. We managed to reconstitute U2 snRNA binding to a model BS in vitro as well as the remodelling leading to the dissociation of HTATSF1 from the complex. We determined a series of cryo-EM structures of the U2 snRNP in different conformational states, including 2 previously unknown assembly intermediates. Our high-resolution reconstructions (2.0-2.3Å) provide unprecedented insight into the architecture and dynamics of the human U2 snRNP and the complex A formation. These new data reveal critical roles of HTATSF1 and SF3B6 in facilitating pre-mRNA binding by the U2 snRNP.
U12-dependent splicing in human cells
In human cells, most introns are processed by the canonical U2-dependent, major spliceosome. However, around 0.5% of human introns utilise an alternative splicing pathway, catalysed by the minor spliceosome, which depends on the U12 snRNA. While U12-dependent introns are rare, they are often located in genes with critical cellular functions, and mutations in the minor spliceosome components lead to several genetic disorders.
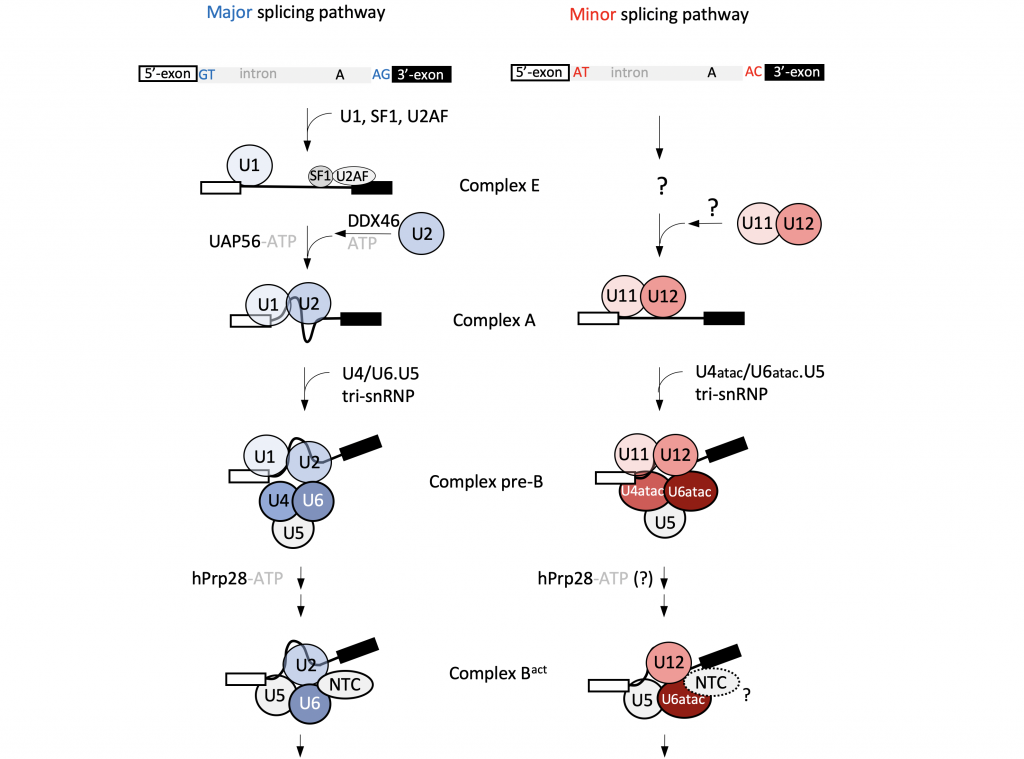
By combining cell-based assays, proteomics, and next-generation sequencing methods, we aim to obtain a comprehensive picture of the minor spliceosome composition and associated regulatory mechanisms.The core of our expertise lies in structural studies by single-particle cryo-EM. We expect that high-resolution structural information for the minor spliceosome will answer some of the fundamental mechanistic questions regarding its assembly and U12-dependent intron recognition. Additionally, it will shed light on structural similarities and differences between the major and minor splicing pathways, and will provide insights into the evolution of the splicing machinery.