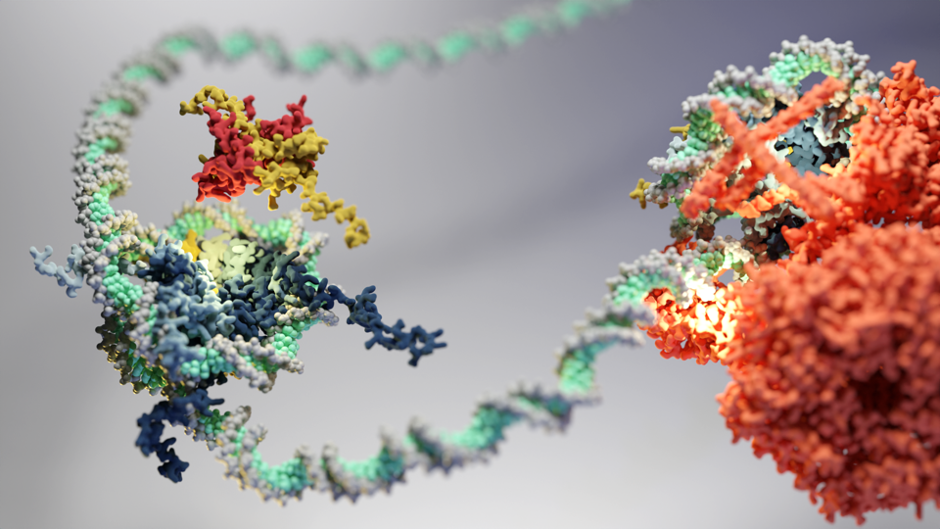
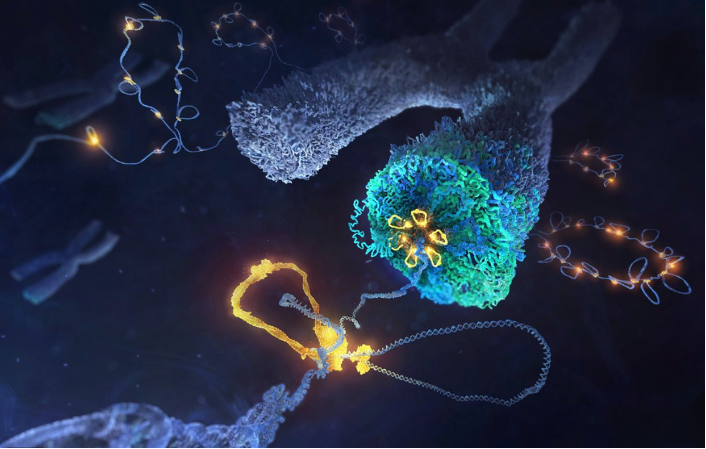
The Eustermann group explores the molecular landscape of chromatin to understand at an atomic level the principles underlying expression and maintenance of genomic information in eukaryotes.

Exploring the chromatin landscape by cryo-electron microscopy

Inside the nucleus of a eukaryotic cell, genomic DNA, proteins and RNA are packaged into a highly condensed, supra-molecular assembly known as chromatin. Given its fundamental role in the control of gene expression and genome maintenance, we are interested in the molecular machinery that actively reconfigures the composition and 3D architecture of chromatin.
The nucleosome is the basic subunit of chromatin, comprising approximately 147 bp of DNA wrapped around a histone octamer. Since its discovery and first structural analysis, fascinating insights into chromatin have been derived from advances in DNA sequencing-based mapping and light microscopy imaging. The genome of a typical human cell, for example, is tightly packed with an estimated 22 million nucleosomes. Coordinated changes in their organisation regulate access to the genetic information and have thus been proposed to control gene expression, by which cells differentiate and maintain their identity. Aberrant nucleosome organisation is closely linked to human diseases such as cancer.
How the tertiary fold and structural dynamics of chromatin underlies its fundamental function remains largely unexplored at a molecular level, even in light of progress in the post-genomic era. Atomic structures have started to emerge that show interactions between individual chromatin factors and parts of the transcriptional machinery. However, the cooperative interplay of such factors in concert with the precise 3D-packing arrangement of nucleosomes, across the vast sequence space of the genome, presents an outstanding challenge for modern structural molecular biology. Our research tackles this challenge by developing system-based approaches to reconstitute the chromosomal landscape in order to visualise its atomic structure and dynamics by advanced methods in cryo-electron microscopy (cryo-EM).
Recently, we determined by single-particle cryo-EM the first high-resolution structure of a multi-subunit ATP-dependent chromatin remodeller and revealed unifying principles by which these enigmatic, megadalton molecular machines govern the genomic position and composition of nucleosomes. In a close collaboration with the Korber group (LMU Munich), we are currently exploiting this remarkable activity to establish whole genome reconstitutions. Rebuilding the chromosomal landscape, including hallmark features around gene promoter regions, enables us to directly probe the principles by which its tertiary structure is generated and reconfigured for transcriptional control, while also providing a powerful approach towards structural studies of the underlying mechanism.
*Readers may view, browse, and/or download this image for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this image may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.