Part 5: Structural analysis
Overview
Congratulations on reaching the final part of our bioinformatics adventure! Thanks to your help we now know that the rhodopsin might be from a new species that is closely related to Sardina pilchardus, a fish that is found along the European coastline. The TREC researchers are very impressed by your skills and have one last favour. As part of their research, they would like to know more about the structure of this rhodopsin.
In this part of the activity, we will explore the structural aspects of proteins using a protein database. The rhodopsin structure of this newly discovered rhodopsin is not available yet. However, we can explore the structure of the closely related Sardina rhodopsin, which should be very similar.
We will focus on the rhodopsin from the Sardina species and use the UniProt (Universal Protein resource) database to examine its secondary structure, specifically alpha helices and beta sheets. Furthermore, we will access the three-dimensional structure of the Sardina rhodopsin using the AlphaFold prediction integrated into UniProt. This will allow us to make reasonable predictions about the structure of the new rhodopsin.
Your task
Please follow the steps outlined below:
1. Search for the Sardina rhodopsin in the UniProt database to find the protein’s secondary structure.
2. Investigate the three-dimensional structure of the Sardina rhodopsin that is created by AlphaFold and displayed in the UniProt database.
3. Try to answer the questions in the “Questions” tab.
UniProt and AlphaFold
1. Access the UniProt database in the window below.
2. Search for the Sardina rhodopsin protein using its UniProt name (OPSD_SARPI) or accession number (Q9YGZ0).
3. On the results page, you will find a summary of all scientific information available about Sardina rhodopsin. Explore the information provided by the UniProt database and try to answer some of the task questions.
4. On the UniProt database window, scroll down to “Structure”. If the structure is not displayed, click on “Click on load the structure viewer”. You are now viewing a three-dimensional structure of the Sardina rhodopsin, also known as protein tertiary structure, that was created by AlphaFold. You can use your cursor to rotate the structure. Additionally, you can click on individual amino acids within the structure to get further information.
5. Have a look at the three-dimensional structure and try to answer some of the task questions.
Questions
1. Based on the information provided by the UniProt database, can you answer the following questions?
- How many transmembrane domains does the Sardina pilchardus rhodopsin have and how are they distributed across the protein? You can find this information in the “Subcellular Location” section.
- Based on the answer to the question before, can you infer the larger group of evolutionarily related proteins to which our rhodopsin belongs?
- What are some common post-translational modifications (PTM) of the Sardina pilchardus rhodopsin? You can find this information in the “PTM/Processing” section.
2. Based on the three-dimensional structure predicted by AlphaFold, can you answer some of the following questions?
- What is the overall shape of Sardina pilchardus rhodopsin?
- Try to identify the individual alpha helices and beta sheets within the structure. The beta sheets are important for the binding of the retina. Where are the two beta sheets located?
Wrap up
Thanks to your help the TREC team could identify the function and evolutionary origins of the unknown protein. Additionally, your results suggest that this rhodopsin might belong to a new species that is closely related to the Sardina pilchardus and lives along the European coastline.
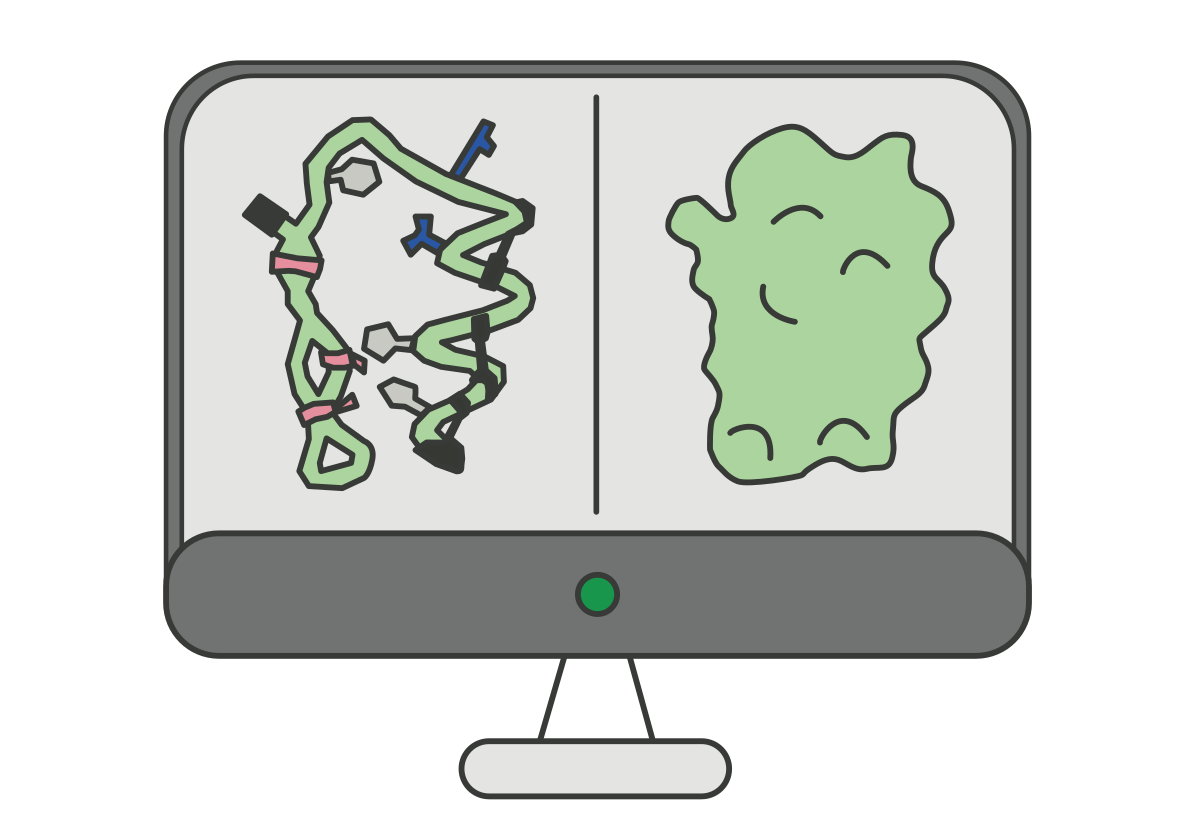
A few months have passed since you helped the team and now, surprisingly, they sent you another email! They explain how they continued their research on this new rhodopsin and with the help of the imaging centre at EMBL, they were able to identify the rhodopsin protein structure from the new species. They used a method called cryogenic electron microscopy (cryo-EM) and they want you to be the first to see the rhodopsin structure. Here it is:
An answer sheet with sample answers for all questions part of the resource is available upon request. Please contact us at schoolvisits@embl.de and make sure to specify this particular resource in your email.
If you want to learn more about bioinformatics please check out this comprehensive set of online resources from EMBL-EBI: https://www.ebi.ac.uk/training/online/courses/introductory-bioinformatics-pathway/.
Activity navigation
Share:
 Deutsch
Deutsch