Part 2: Protein identity and function
Overview
In Part 1, we successfully translated the unknown DNA sequence to an amino acid sequence. However, our expedition to unravel the mysteries of this unknown protein has only just begun. At this stage, we are still unaware of its function. To gain further insights, we will explore a comprehensive database of known protein sequences to identify any similar sequences that might shed light on the function of our protein.
We will use the bioinformatic tool NCBI BLAST+ to assist us in this process. NCBI BLAST+ enables us to identify and compare amino acid sequences and here, we will use it to identify known sequences that are similar to our unknown sequence.
Your task
Please follow the steps outlined below:
1. Copy the amino acid sequence that you have recently translated. You can find it in the previous task or download it from the provided link here. Use a text editor tool (e.g. TextEdit and Notepad) to open FASTA files.
2. Proceed to the “NCBI BLAST+” tab and carefully follow the instructions provided to search for the identity of the protein using the NCBI BLAST+ tool.
3. Try to answer the questions in the “Questions” tab.
NCBI BLAST+
1. Access the NCBI BLAST+ tool in the window below.
2. In STEP 1, ensure that the database ”UniProtKB/Swiss-Prot” is selected.
3. In STEP 2, paste your amino acid sequence in the query box.
4. In STEP 3, ensure the program “blastp” is selected.
5. Run the search by clicking on “Submit”.
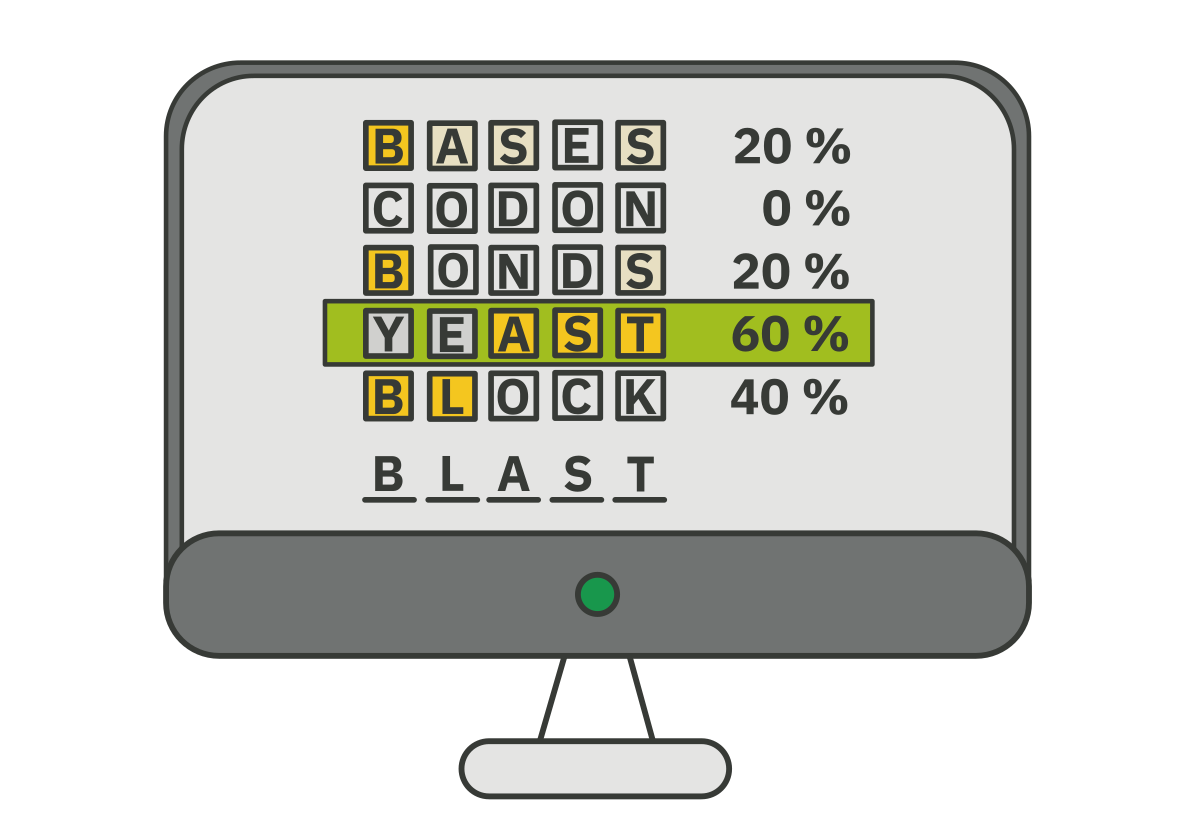
6. Examine the data table (“Summary Table”), and try to answer the questions related to the task.
Note: The summary table displays sequences from the database that are listed according to their similarity to the inserted sequence. The most similar sequence, also known as the best hit, is displayed at the top of the list.
Questions
1. Which protein and species is the best hit in the protein database?
2. What is the percentage of identity shared by the “unknown protein” and the best hit?
3. What is the known biological function of the best hit? To get this information, right-click on the second column of the results table (DB:ID) and open the link in a new tab.
Activity navigation
Share:
 Deutsch
Deutsch