Bateman Group
EMBL
Creating synergies between EMBL and Stanford’s research communities
Saccharomyces cerevisiae is a useful model for the understanding the basis of cellular biology including chromosome maintenance and the cell cycle. S. cerevisiae can inform genetic medicine via annotation of human disease-related phenotypes and gene function due to functional complementation between yeast and human homologs. The budding yeast is the most well studied eukaryotic genome which has led to a vast range of the experimental literature.
The Saccharomyces Genome Database (SGD) is the comprehensive resource containing the highest quality curated information about the S. cerevisiae genome and its elements. The SGD provides a comprehensive resource that facilitates experimentation in other systems. It annotates the genome, assimilates new data, includes genomic information from other fungal species, and incorporates formalized and controlled vocabularies to represent biological concepts. Databases such as SGD provides a substantial service to the scientific community, reaching out to researchers in the greater biomedical research community to serve those who have a need for information about genes, their products, and their functions.
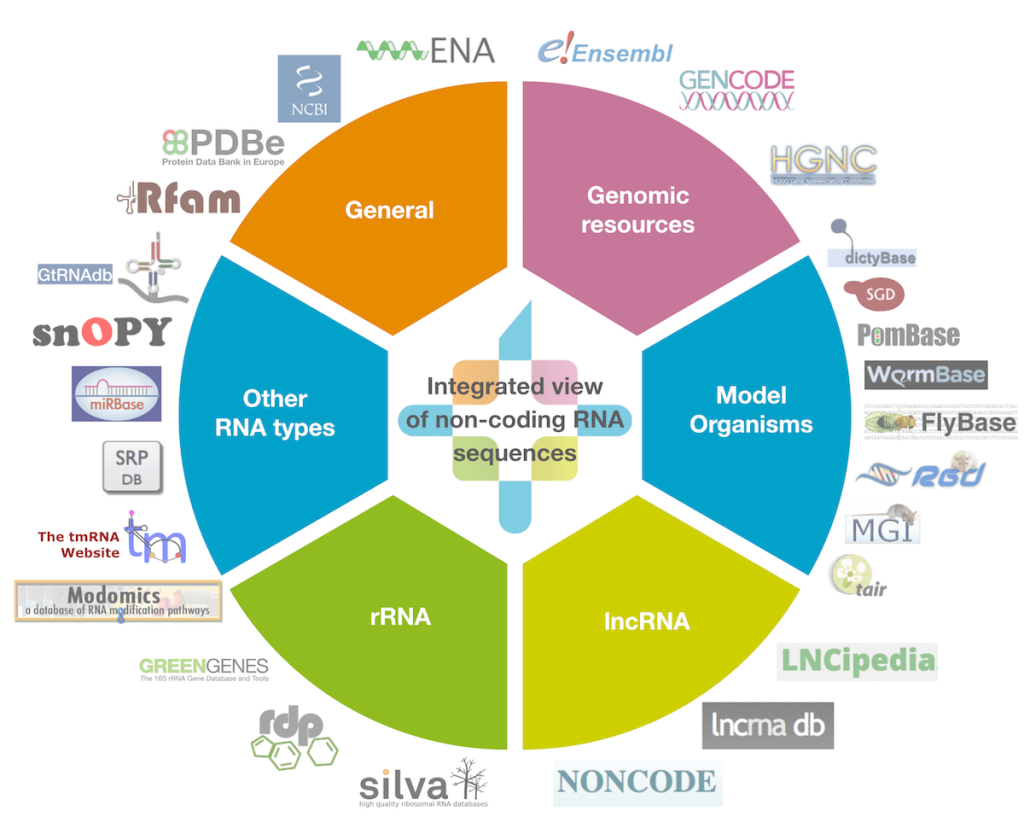
Non-coding RNAs (ncRNAs) are a critical component of cellular machinery of all organisms. There is an intense scientific interest in ncRNAs resulting in a large number of ncRNA databases, but until recently searching and comparing them was challenging, and there was no uniform way to access or reference ncRNA sequences.
In collaboration with RNAcentral we provide gene models and annotations for the non-coding RNAs found in the S. cerevisiae reference genome sequence. The ncRNAs found within other genomes from this species will be provided as annotations are created. Providing a comprehensive, centralized database for ncRNAs facilitates the next generation of RNA research and help drive further discoveries.
Publications:
The Kipoi repository accelerates community exchange and reuse of predictive models for genomics. Z Avsec, R Kreuzhuber, J Israeli, N Xu, J Cheng, A Shrikumar, A Banerjee, DS Kim, T Beier, L Urban, A Kundaje, O Stegle, JGagneur. Nature biotechnology (2019).
Interested in finding out more about the ncRNA annotation project? Do you want to contribute this great resource for scientists? Get in touch, we would love to hear from you!

EMBL

Stanford
RNAcentral: a hub of information for non-coding RNA sequences
The RNAcentral Consortium, RNAcentral: a hub of information for non-coding RNA sequences, Nucleic Acids Research, Volume 47, Issue D1, 08 January 2019, Pages D221–D229