eSPC: a user-friendly platform for analysing biophysical data
ARISE fellow Osvaldo Burastero is developing eSPC, an interactive open-source platform for biophysical data analysis
Characterising the homogeneity, stability, conformation, and interactions of biomolecules is fundamental to the understanding of all biological processes. To study these properties, scientists employ a multitude of well-established biophysical methods, including differential scanning fluorimetry, dynamic light scattering, microscale thermophoresis, circular dichroism and biolayer interferometry, among others. The downstream analysis of these experiments can be time-consuming and challenging, especially for scientists without coding skills.
The EMBL Sample Preparation and Characterization (eSPC) platform, developed by Osvaldo Burastero and his collaborators, alleviates this burden by providing easy-to-use software for the analysis of biophysical data.
Osvaldo earned his PhD in Chemical Biology and an MSc degree in Data Mining and Knowledge Discovery at the University of Buenos Aires, Argentina. As an ARISE fellow in Maria Garcia Alai’s team at EMBL Hamburg, he integrates both biophysical and computational expertise to advance the eSPC platform.
What are the main features of the eSPC platform?
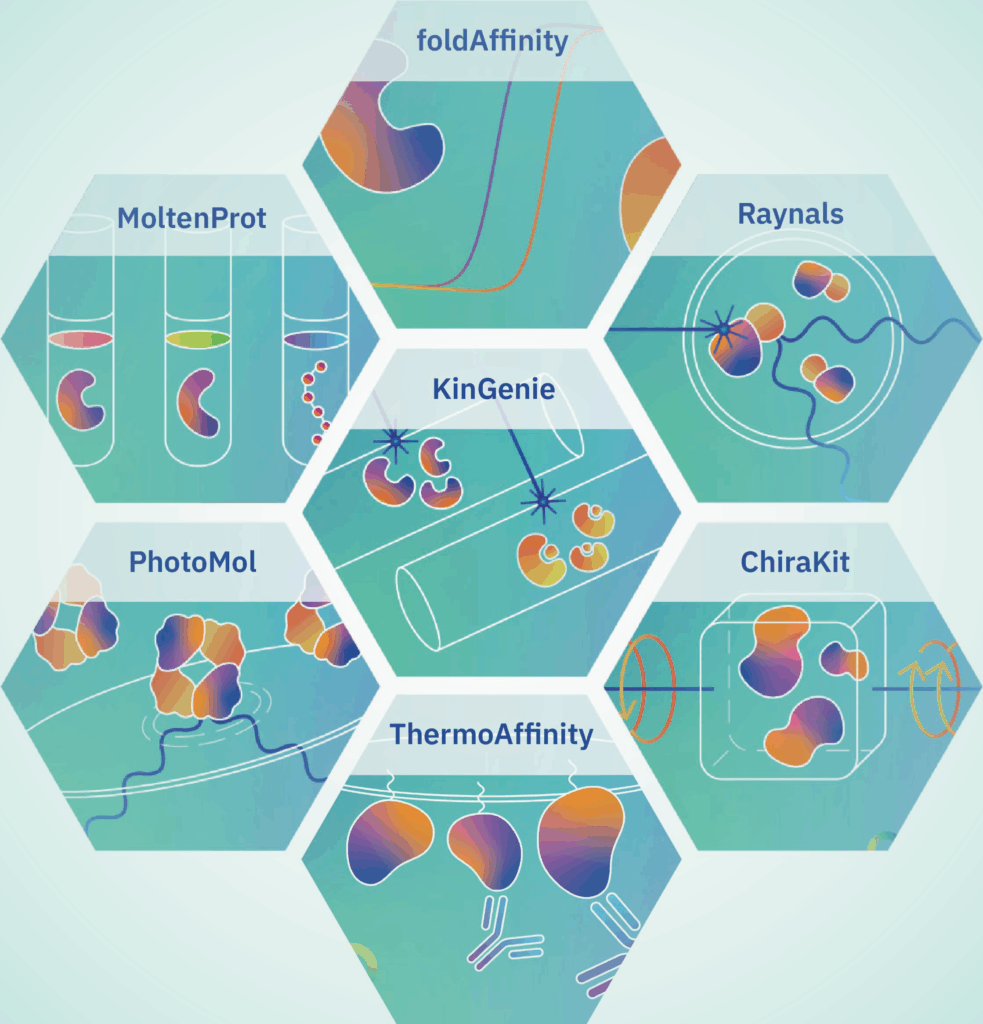
The eSPC platform provides interactive tools for analysing the properties of biomolecules, including homogeneity, size, mass, stability, and interactions. Currently, the platform offers seven tools, which are described below.
MoltenProt: For studying protein stability. It can analyse protein thermal denaturation data obtained by differential scanning fluorimetry.
FoldAffinity: For determining binding affinities when ligand binding stabilizes the protein.
ThermoAffinity: For binding affinity studies using microscale thermophoresis and steady-state spectroscopy. It can estimate equilibrium dissociation constants when ligand binding produces a shift in the thermophoresis or fluorescence.
ChiraKit: To analyse circular dichroism data. It includes submodules for determining secondary structures, fitting thermal and chemical unfolding models, evaluating peptide alpha-helix content, and performing spectral comparisons (e.g., for quality control).
Raynals: To analyse particle size and homogeneity in the nanometer range using data obtained by dynamic light scattering.
PhotoMol: For assessing the mass of molecules in solution using mass photometry data. It is useful to know a protein’s oligomeric state.
KinGenie: For the analysis of interaction kinetic data obtained by biolayer interferometry. It can estimate on- and off-rates of binding.
What advantages does the eSPC platform offer for the analysis of biophysical experiments?
One advantage is that the tools are open-source, so the software does not work as a ‘black-box’ and users can inspect the code to understand how the data is processed and fitted. Then, the eSPC is online, so there is no need for installation or analysing the data in the same machine where the experiment was performed. On top of it, the tools are interactive, so users can visualise their data and immediately assess the effect of applying various models with different parameters.
Moreover, all the tools within the platform share the same interface, making it easy for users to navigate from one tool to another once they become familiar with a single one. We also provide tutorials and example datasets to assist users in their data analysis. Additionally, eSPC offers unique analysis features that are not available in any other software.
What aspects of ARISE have contributed to your project’s progress?
The ARISE programme offers excellent training opportunities, including secondments with partner organisations in the private or academic sectors. During my fellowship, I completed my secondments at the Institut Pasteur and Refeyn.
At the Institut Pasteur, I worked under the supervision of Patrick England. This experience strengthened my biophysical data analysis skills and allowed me to receive expert feedback on the eSPC tools. I also obtained hands-on training with biophysical instruments, which was crucial for deepening my understanding of the techniques before developing the software. Last, I could promote the eSPC platform to a wider audience and gain more users.
During my short industry secondment at Refeyn, I worked under the supervision of Max Hankte. Here, I shadowed the Software Team and learnt from their software development good practices, which will make the eSPC easier to maintain in the future. This secondment was my very first experience in industry, and it opened up alternative career paths for me.
What is on the horizon for the eSPC platform?
We hope to finish the development of KinGenie in the coming months. Additionally, we are planning to release a new tool for the global analysis of thermal and chemical denaturation data. Furthermore, we are experimenting with AI and the implementation of a chatbot for the analysis of biophysical data.
What hobbies or interests do you enjoy outside of work?
I enjoy practicing martial arts, such as Jiu-jitsu and Judo, as well as learning languages. I love spending time with my partner and family, and if possible, outdoors. Lately, I have been learning how to use AI for analysing data. Hopefully, I can apply it to the eSPC too.