Read the latest Issue
By combining three different kinds of microscopy, scientists at EMBL Heidelberg have been able to determine, for the first time, how cellular machines move in relation to each other as the cell’s membrane bends inwards to form a vesicle that carries material into the cell.
Whether it’s a white blood cell engulfing a bacterial invader or a microbe gobbling up supper, cells often take in large items by bending their own membrane around them and folding it inwards, like poking in a finger on a glove. This seemingly mundane process involves around 50 different molecules, and Marko Kaksonen’s lab at EMBL Heidelberg is set on untangling just how they do it.

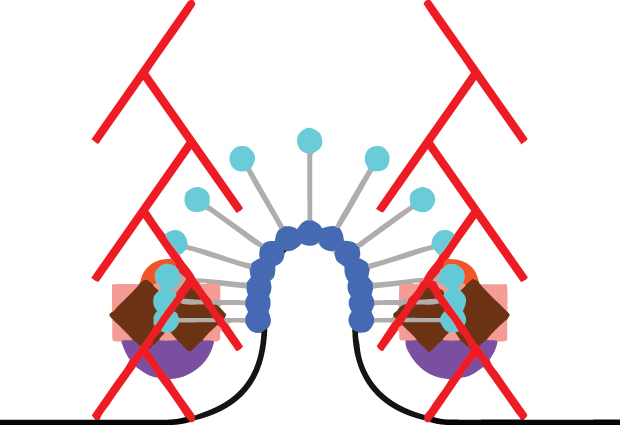
“It’s actually not a cartoon, it’s a reconstruction,” says Andrea Picco of the animation above. “We know the number of molecules, and we know where the centre of mass of the structure they form is. With our approach we can measure and track the movement of those molecules in relation to each other, and we can map that onto the shape of the membrane at each point in time, to really reconstruct how things unfold.”
To carry out this reconstruction, Picco, a postdoc in the Kaksonen and Nédélec labs, combined the best of three worlds. He developed a method which enabled him to track the average positions of the different molecules using conventional light microscopy – “electron microscopy can’t follow movement, and super-resolution microscopy is still too slow for this,” says Kaksonen. Picco then plotted those molecular paths onto a map of the cell: detailed information on the changes in the shape of the cell membrane as it folds inwards, obtained by electron microscopy. To delve beyond this map of light trails to a more detailed understanding of individual molecules’ structure, Picco teamed up with Markus Mund from Jonas Ries’ lab, which specialises in super-resolution microscopy. Thanks to this added degree of precision, the scientists discovered, for instance, that a protein called Sla1 (the purple circles in the animation) forms a ring around the tip of the membrane ‘finger’.
To the expert eye, this animation also reveals the answer to a puzzle: which direction do actin filaments (the red rod-like elements in the animation) grow in? This has been a debated topic, with different scientists putting forth different possibilities. But by letting the actin network form, then turning off the fluorescent tags on it, and watching where new tags come back on, Picco and Kaksonen were able to show that new actin molecules are added at the base, and the filaments all grow inwards – likely pulling the membrane with them.
“We’re beginning to go beyond just a description of the structure, to get to the actual function,” says Kaksonen. His lab now plans to go further in that direction, by tampering with the different machines and analysing how the whole membrane-bending process is affected.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive