
Read the latest Issue
Ernst Stelzer earns 2016 Lennart Philipson award for advances in light sheet microscopy
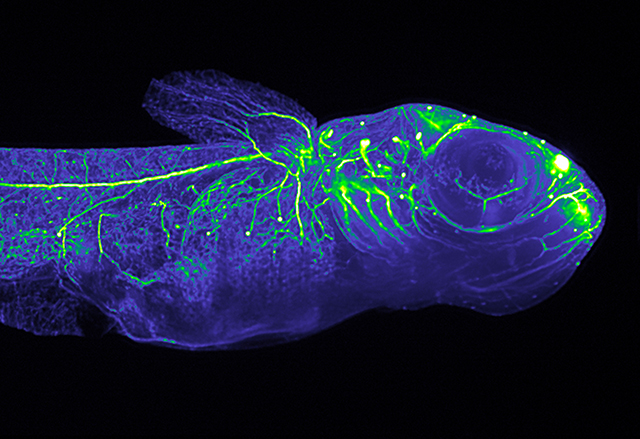
For Ernst Stelzer, coming back to EMBL’s Heidelberg campus “is like coming back to an old friend.” He arrived in the ‘1980s’ as a physicist, and the innovative technologies developed by his teams have paved the way for biologists to further understand life at the microscopic level, and in unprecedented detail – from how neurons communicate with one another, to monitoring the beating heart of a fish. In recognition of his contributions, Stelzer was awarded this year’s Lennart Philipson award by the EMBL Alumni Association.
“The University of Frankfurt, where I am now based, may have been my alma mater,” Stelzer says, “but let’s face it: I spent 28 years here at EMBL.” Among the first group of PhD students, Stelzer began at EMBL’s Heidelberg site in 1983 working on confocal fluorescence microscopy. The atmosphere at EMBL was quite different back then. For one thing, the lab did not possess a single fluorescence microscope. Even then, “in those days, it was not yet thought of as a quantitative tool or measuring device,” Stelzer says. “Microscopy was more thought of as a way to just visualise something.”
By 1993, together with his postdoc Stefan Hell, Stelzer had developed 4Pi microscopy. During the same year he and his PhD student Steffen Lindek patented and built confocal theta microscopes, a project that later involved James Swoger. By 1998 the team started to build something called a tetrahedral microscope, a machine that improved resolution using four lenses. Every calculation and equation they did over the years, though, brought them closer and closer to a striking revelation. The team realised that they could build a machine that would let them see life as never before. They set out in search of spare parts around the lab – an unused lens, a laser tucked away in a closet, a motor abandoned by a former postdoc. From these, they constructed an early version of a remarkable machine, one of the first implementations of light-sheet based fluorescence microscopy.
The difference between traditional microscopy techniques and the machine Stelzer built can be illustrated using the example of imaging a developing fruit fly embryo. Previous microscopes would shine light evenly on the fly egg. And because its yolk is opaque, light would scatter and make it very difficult to see inside – let alone visualise it in 3D. Additionally, the amount of light used in the system can expose the specimen to up to 1000 times more energy than we typically receive from the sun. “You would not want to go to the beach in that!” Stelzer jokes.
We don’t just look in one direction. We look at eight different directions in order to get a whole view of the specimen.
The new microscope overcame both of these obstacles. First, rather than repeatedly illuminating the entire fly – and frying it to a crisp – their microscope shone light through the egg in very thin slices, or planes. “Imaging in 3D is different than 2D,” Stelzer says. “We don’t just look in one direction. We look at eight different directions in order to get a whole view of the specimen.” Scientists can then put these slices together using a computer and digitally recreate the specimen in 3D. And since less light energy is being used to image the egg at a time, the fly can survive imaging for days at a time. Selective plane illumination microscopy, or SPIM, was born.
The next challenge was to convince other biologists. “People seemed hesitant to try something so new at the time,” Stelzer remembers. But EMBL alumnus Jochen Wittbrodt, who was working with medaka fish, stepped up to the challenge. They brought on other students, including Sebastian Enders, Jan Huisken and Philipp Keller. Together, they did something no scientist had done before: see the inside of a live, intact fruit fly embryo, and image it in 3D.
The results were astounding. Through thousands of images put together like a detailed flipbook, the team could clearly see and trace the organs as they developed inside the fly embryo – the pinching off of thorax and abdomen, the banding of rounded segments, the growth of the eyes. The whole period of imaging lasted seventeen hours. And what’s more, the fly survived intact and was able to complete embryogenesis. Without delay, Stelzer and Steffen Lindek wrote a patent for SPIM and the team got to work on a prototype.
Life is not flat – cells don’t grow between a glass slide and cover slip. Cells grow on cells!
By the early 2000s what began as a side project became a full-time focus. The team had constructed a monolithic microscope carved out of a single block of aluminium. Later versions were built in increments with increasingly advanced lenses – and they even turned to German car manufacturers for other parts. “The motors that we ordered from one company were not even in the catalogue yet!” Stelzer remembers.
Applications of light-sheet based fluorescence microscopy have come a long way since this adventurous group of scientists were hacking parts together to think in 3D. Researchers can now use it to probe into deep tissue, such as in plants and zebrafish, as well as better understand diseases that affect humans, such as type 1 diabetes and congenital disorders. “You take on a different attitude toward your specimen when thinking in 3D,” Stelzer says. “Life is not flat – cells don’t grow between a glass slide and cover slip. Cells grow on cells! The seeds of light-sheet based fluorescence microscopy were sown at EMBL – and thanks to the efforts of biologists, physicists, engineers and many other professionals, its applications are now being developed and used around the world.”
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive